Clinical impact of oncomirs 221 and 222 on breast cancer diagnosis
Elham Ali Ahmed 1, Sohair A. Abd El-bast1, Mona A. Mohamed2, Menha Swellam3,4
1Zoology Department, Faculty of Science (Girls), Al-Azhar University.
2Biochemistry Division, Chemistry Department, Faculty of Science (Girls), Al-Azhar University.
3Biochemistry Department, Genetic Engineering and Biotechnology Research Division, AI-Azhar University.
4High Throughput Molecular and Genetic laboratory, Center for Excellences for Advanced Sciences, National Research Centre, Dokki, Giza, Egypt.
Correspondence: Professor Dr. Menha Swellam ( High Throughput Molecular and Genetic laboratory, Center for Excellences for Advanced Sciences, Biochemistry Department, Genetic Engineering and Biotechnology Research Division, National Research Centre, El-BohouthStreet, Dokki, Giza, 12622, Egypt.; E-mail: menhamswellam@gmail.com).
Asia-Pacific Journal of Oncology 2020, 1: 9-17. https://doi.org/10.32948/ajo.2020.07.18
Received: 04 Nov 2019 | Accepted: 07 Jan 2019 | Published online: 26 July 2020
Background Dysregulation of miRNAs, non-coding RNAs of 18-25 ( ̴ 22nt), is a hallmark of malignancies among them is breast cancer. The present study aimed to investigate the expression levels of circulating oncomiRNAs (miRNA-221and miRNA-222) as a minimally non-invasive method for early detection of breast cancer as compared to tumor markers (CEA, CA15.3).
Materials and methods MiRNA-221 and miRNA-222 expression levels were determined using quantitative real-time polymerase chain reaction (qPCR) in serum samples from three groups: primary breast cancer patients (n = 44), benign breast lesion patients (n = 25), and healthy individuals as control group (n = 19). Their diagnostic efficacy and relation with clinicopathological data were analyzed.
Results MiRNA-221 and miRNA-222 expression and tumor markers reported significant increase in their mean levels in breast cancer group as compared to the benign breast lesions or control individuals. Among clinicopathological factors, miRs reported significant relation with pathological types, clinical staging, histological grading and hormonal status, while CEA and CA15.3 did not revealed significance with these factors. The diagnostic efficacy for investigated miRNAs was superior to tumor markers especially for detection of early stages and low grade tumors.
Conclusion MiRNA-221 and miRNA-222 were superior over tumor markers for early detection of breast cancer especially those at high risk as primarybreast cancer patients with early stage or low grade tumors.
Key words Breast cancer; miRNAs; circulating molecular marker; diagnosis
Early detection of breast cancer greatly improves the prognosis and treatment for patients, this critical for optimizing management strategies to this disease, but in spite of improvements in cancer screening techniques, major challenges remain [5, 6]. Cancer antigens (such as CEA and CA15.3) as well as other circulating protein molecules have been widely used as non-invasive biomarkers for monitoring patients with metastasis, but with insufficient sensitivities to diagnose primary breast cancer [7, 8]. Thus, great necessity needed to develop novel biomarkers for breast cancer detection with high sensitivity and specificity.
MicroRNAs (miRNAs) are a family of highly conserved noncoding single stranded RNA molecules of 21 to 25 nucleotides; It is can regulate one third of protein-coding genes and participate in the developmental and physiological processes of human body [9]. Aberrant expression of miRNAs has a link with breast tumorgenesis and their deregulation is reported in biological fluids of cancer patients [10]. MiRNAs have important regulatory roles in the cell; modulate their target genes through RNA interference pathways (RNAi). It can reversely modulate gene expression processes [11]. Circulating miRNAs are stable in body fluids, they have been expressed in many cancers among them is breast cancer, miRNAs are potential bio-markers in the diagnosis, prognosis, and prediction of response to treatment in breast cancer [12, 13].
In the current study, miRNA-221 and miRNA-222 are homologous miRNAs located in tandem, within 1 kb from each other, on human X chromosome [14, 15]. Recent studies stated that microRNA-221 and miRNA-222 is aberrantly expressed in various malignancies [16] and was found to be overexpressed in triple-negative breast cancer patients [17]. MiRNA-221 and miRNA-222 are thought to serve as oncomiRs because they inhibit many tumor suppressors [18, 19, 20, 21].
The present study aimed to investigate the potential diagnostic role of circulating miRNA-221and miRNA-222 in blood samples from breast cancer patients as minimally invasive method and correlate their results with both tumor markers and clinicopathological data.
After obtaining ethical approval from the Scientific Medical Ethical Committee (National Research Centre, number#15029), and between February 2018 and January 2019 a total of 88 females were enrolled in the current study and they were categorized into three groups based on their diagnostic criteria: primary breast cancer group (n=44), female patients with benign lesions (n=25) and healthy individuals (n=19) served as control group. All groups were of matched ages. The enrolled individuals who fulfilled the inclusion criteria (newly diagnosed breast cancer patients, and before they start any treatment strategies) signed their informed consent, while those with distant metastasis or who received radio-or- chemotherapy were excluded from the study. Clinicopathological data for enrolled individuals were collected from their clinical sheets, and breast cancer patients were diagnosed according to their staging and grading systems following the TNM classification [22] and modified Scarff-Bloom Richardson histologic grading system [23], respectively. The expression of hormonal receptors was examined using immunohistochemistry method as previously reported [24]. Both ER and PgR were consider positive if ≥10%nuclei was positively stained using 10 high-power field, andHER-2neu were considered positive if scored as +3 [25].
Sample collection and processing
Three milliliter blood samples were collected from all enrolled individuals in tubes with polymer gel and clot activator (Greiner bio-one, GmbH, Australia) then allowed to clot at room temperature for 30min, and all samples were centrifuged at10,000×g for 10min at 4°C (13-18KS, Sigma, Germany). Separated sera were aliquoted and stored at −80 °C for miRNA expression analysis.
Assessment of Tumor markers
By using enzyme-linked immunosorbent assay (ELISA) both tumor markers, CEA and CA15.3 were detected in serum samples using available commercial ELISA kit (Immunospec Corporation, Canoga Park, CA, USA). According to instructions in the manual manufacturer’s protocol, their concentration was detected using GloMax®-Multi detection system (Promega, Fitchburg, WI, USA).
MiRNA extraction
Circulating miRNA was extracted from the serum samples using miRNeasy Mini kit (Catalogue # 217004, Qiagen, USA), as recommended by the manufacturer's instructions, briefly: QIAzol Lysis Reagent (RNA extraction reagent) was added to serum samples (with volume ratio 5:1) and then vortex was applied. After incubation for 5 min at room temperature (25 °C) Lysates were left to promote dissociation of nucleoprotein complexes. Phase separation step was carried out by adding an equal volume of chloroform to the tube containing the lysate, and vortex followed by centrifugation for 15 min at 12,000 xg at 4 °C using cooling centrifuge (13-18KS, Sigma, Germany). The upper aqueous phase was transferred to a new collection tube. Then 1.5 volumes of 100% ethanol were added to the aqueous phase followed by pipetting up and down several times. Up to 700 μl of the sample was transferred into an RNeasy Minispin column (Qiagen) in a 2 ml collection tube and centrifuged for 15 s at ≥ 8000×g for 15 min at room temperature. The RWT buffer (700 μl) was added to the RNeasy Mini spin column the centrifugation was applied and the flow-through was discarded. Afterwards, 500 μl of RPE buffer was added then the column was centrifuged and flow-through was discarded, this step was repeated. The RNeasy Mini spin column was placed into a new 2 ml collection tube and centrifuged at full speed for 2 min. After washing steps; the RNeasy Mini spin column was placed into a new 2 ml collection tube and centrifuged at full speed for 2 min. Finally, the RNeasy Mini spin column was transferred to a new 1.5 ml collection tube. The RNA was eluted using RNase-free water in a final volume (30 μl) was added directly onto the RNeasy Mini-spin column membrane and centrifugation was done for 1min at≥8000×g. The purity and the concentration of the purified miRNA was detected using nano-drop spectrophotometer (Quawell, Q-500, Scribner, USA) and stored at −80 °C till further assessments.
Reverse transcription and cDNA preparation
Reverse transcription of miRNA was performed MiScript II reverse transcription kit (Cat number # 218160, Qiagen, USA).As recommended in the manufacturer's instructions by using a total volume of 20 μl of reverse transcription reaction components as follows: 4 μl MiScript HiFlex buffer, 2 μl nucleic mixture, 2 μl MiScript RT mixture, and RNase-free water (variable depending on the volume of the added template miRNA) and template of purified miRNA with adjusted concentration 100 ng/ml. The polymerase chain reaction (PCR) tubes were then placed in thermal cycler (Sure Cycler 8800, Agilent, USA) and the transcription profile was adjusted for 60min at 37 °C. Complementary DNA concentration and purity were detected using nano-drop spectrophotometer (Quawell, Q-500, Scribner, USA) and stored at −20 °C till performing qPCR.
Quantitative real-time PCR (qPCR)
Quantitative real-time PCR was carried out using miScript primer assay (Cat number 218300, Qiagen, USA) for miRNA-221 (Hs_miR_221_2miScript Primer Assay, MS00009079), miRNA-222 (Hs_miR_222_2miScript Primer Assay, MS0007609), the reaction was performed using MiScript SYBR Green PCR kit (Cat number 218073, Qiagen, USA).Also, RNU6–2 (Hs_RNU6-2_11 miScript Primer Assay, MS00033740) was used as an endogenous control to normalize the expression levels of the investigated miRNAs. The reaction for miScript primer assays were carried out by using cDNA with concentration adjusted to 2 ng/ml and atotal volume of 20 μl, whereas thermal reaction conditions were as follows: 95 °C for 15min followed by 40 cycles at 94 °C for 15 s, 55 °C for 30 s, and 70 °C for 34 s, in which fluorescence was acquired and detected by Strata gene Real-time PCR system (Max3005P QPCR system, Strata gene, Agilent biotechnology, USA). The expression levels of the investigated miRNAs were evaluated using the ΔCt method 26. The cycle threshold (Ct) value is the number of qPCR cycles required for the fluorescent signal to cross a specified threshold. ΔCt was calculated by subtracting the Ct values of RNU6–2 from those of investigated miRNAs. ΔΔCt was calculated by subtracting the ΔCt of the control samples from the ΔCt of the cancer samples.
Statistical analysis
The fold change values in investigated miRNAs was calculated using the equation of 2ΔΔCT. The association between the clinicopathological and demographic factors with investigated miRNAs was determined by ANOVA analysis. The optimal cutoff point was determined using Receiver Operating Characteristic (ROC) Curve which was plotted by calculating the true-positive fraction (sensitivity [%]) and false-positive fraction (100- specificity [%]) and the best cutoff point was detected as the point that maximize the sum of the sensitivity and the specificity and area under curve (AUC) approach to 1 (range 0.5-1). Accordingly ROC curbves for investigated items were blotted to detect the sensitivities and the specificities for the miRNAs and tumor markers with their diagnostic efficiencies 27. Correlations between investigated miRNAs were analyzed using the Spearman's correlation coefficient (CC). SPSS (version 10 SPSS, Inc., Chicago USA) was used in data analyzing and P-value were two-tailed and considered significant if < 0.05.
Diagnostic Efficacy of circulating miRNA-221 and miRNA-222
Mean levels of investigated miRNAs reported significant difference among studied groups as the miRNA-221 level was higher in breast cancer group as compared to the other two enrolled groups (benign and healthy control groups) as reported in Table 1, the median level was higher 9.2 folds in breast cancer (median119) compared to benign lesion (median 12.9) and 25.3 folds than healthy control (median 4.7) at P<0.0001, and the benign. Similarly miRNA-222 the mean levels were superior in breast cancer group as compared to benign and control ones as reported in Table 1. The increment in the median levels was 2.7 folds higher in cancer patients (median 52.8) as compared to benign ones (median 19.5) while it was 9.8 folds as compared to control group (median 5.4). Tumor markers CEA and CA15.3 also reported significant increase in breast cancer patients as compared to the other two studied groups as shown in Table 1.
The positivity level was considered as values above the cutoff points which have been reported from the ROC curves as shown in Figure 1 as 32.4 fold change for miRNA-221, 21.6 fold change for miRNA-222, 15.3 ng/ml for CEA and 21 ng/ml for CA15.3. Considering these cutoff points the positivity rates were significantly increased in cancer as compared to benign or control group as reported in Table 1.
Relation between miRNAs , tumor markers and clinicopatholical factors
MiRNA-221 expression level reported significant difference (F=5.9, P=0.019) with pathological types as its level was increased among breast cancer patients with invasive duct carcinoma (IDC) (mean ± SEM) (195.7 ± 26) as compared to those with duct carcinoma in situ (DCI) (99 ± 25). Also miRNA-221 expression level was increased with positive lymph nodes (203.8 ± 29) as compared to its expression (112.7 ± 24) in negative lymph nodes at (F=5.56, P=0.023). Clinical stages reported significant difference with miRNA-221 as for early stage it was (104.5 ± 24), while for late stage it was (195.5 ± 28) at (F=5.34, P=0.026). Histological grading was significantly related with miRNA-221 expression as increased expression was observed with high grade breast cancer (190 ± 26) as compared with low grade (102 ± 26) at (F=4.6, P=0.035). Among hormonal status, ER- status did not report significant level while significant difference was reported between PgR and HER-2/neu positive levels with miRNA-221 as shown in Figure (2 A-B).
MiRNA-222 expression level showed significant relation (F=4.9, P=0.032) with pathological type since its mean level (mean ± SEM) was increased among patients with IDC (134 ±30) as compared to those with DCI (46.2 ±10). For clinical staging the expression of miRNA-222 was significantly (F=4.08, P=0.05) as in early stage it was (52.3 ±11) and for late stage (133.5 ±32). Also its expression was increased significantly (F=6.1, P=0.017) with high grade breast tumor (136.4 ±29) as compared to low grade ones (37.6 ±6.5). Regarding hormonal status, significant difference (P<0.05) was reported between miRNA-222 with PgR and HER-2/neu as shown in Figure (2C-D).
Both CEA and CA15.3 reported no significant relation with clinicopathological factors.
Concordance between investigated markers and enrolled groups
Assessment of investigated markers; miRNAs and tumor marker regarding enrolled individuals (n=88) was reported in Table 2, while among breast cancer group (n=44), significant relation between both miRNAs (R= 0.433, P=0.003) was detected.
Overall sensitivities, specificities, PPVs, NPVs and accuracies of miRNAs and tumor markers among breast cancer diagnosis and high risk groups
As reported in Table 3, the sensitivity and PPV of both miRNA-221 and miRNA-222 was superior over the other investigated markers (CEA and CA15.3) for early detection of breast cancer especially those at high risk as early stage and low grade tumors.
|
Table (1): Level and Positivity rates of investigated markers among the three studied groups. |
||||||||
|
|
MiR-221 |
MiR-222 |
CEA |
CA15.3 |
||||
|
Mean± SEM |
Positivity |
Mean ± SEM |
Positivity |
Mean ± SEM |
Positivity |
Mean ± SEM |
Positivity |
|
|
Control |
8.18 ± 1.7 |
0% |
5.6 ± 0.28 |
0% |
8.86 ± 0.84 |
0% |
12.4 ± 1.3 |
0% |
|
Benign lesion |
21.5 ± 3.3 |
7.7% |
26.6 ± 5 |
18.4% |
12.35± 0.85 |
20.0% |
16.4 ± 1.4 |
32.0% |
|
Breast cancer |
158.3 ± 2 |
92.3% |
100 ± 2 |
81.6% |
13.1 ± 1.1 |
31.8% |
19.7 ± 1.4 |
52.3% |
|
|
F=24, P<0.0001 |
X2= 50, P<0.0001 |
F=8.3, P<0.0001 |
X2= 49, P<0.0001 |
F=3.6, P=0.03 |
X2= 7.98, P=0.018 |
F=5.5, P=0.006 |
X2= 16, P<0.0001 |
|
Table (2): Correlation between investigated markers and enrolled individuals (n=88). |
||||
|
Markers |
MiR-221 |
MiR-222 |
CEA |
CA15.3 |
|
R, P |
R, P |
R, P |
R, P |
|
|
MiR-221 |
--- |
0.652, 0.0001 |
0.275, 0.01 |
0.372, 0.0001 |
|
MiR-222 |
0.652, 0.0001 |
--- |
0.225, 0.017 |
0.301, 0.004 |
|
CEA |
0.275, 0.01 |
0.225, 0.017 |
--- |
0.012, 0.91 |
|
Table (3): Overall sensitivities, specificities, PPVs, NPVs and accuracies of miRs and tumor markers among breast cancer diagnosis and high risk groups. |
|||
|
|
Breast cancer diagnosis |
Early stage |
Low grade |
|
MiR-221 Sen.% Spec% PPV NPV Acc. |
92.3 83.7 81.8 93.2 87.5 |
77.8 93.2 82.4 91.1 88.7 |
81.2 93.2 81.2 93.2 90 |
|
MiR-222 Sen.% Spec% PPV NPV Acc. |
81.6 89.7 90.9 79.5 85.2 |
77.8 79.5 60.9 89.7 79 |
75 79.5 57.1 89.7 78.3 |
|
CEA Sen.%
Spec%
PPV
NPV
Acc. |
31.8
88.6
73.7
56.5
60.2 |
22.2
88.6
44.4
73.6
69.4 |
18.8
88.6
37.5
75
70 |
|
CA15.3 Sen.%
Spec%
PPV
NPV
Acc. |
52.3
81.8
74.2
63.2
67 |
55.6
81.8
55.6
81.8
74.2 |
56.2
81.8
52.9
83.7
75 |
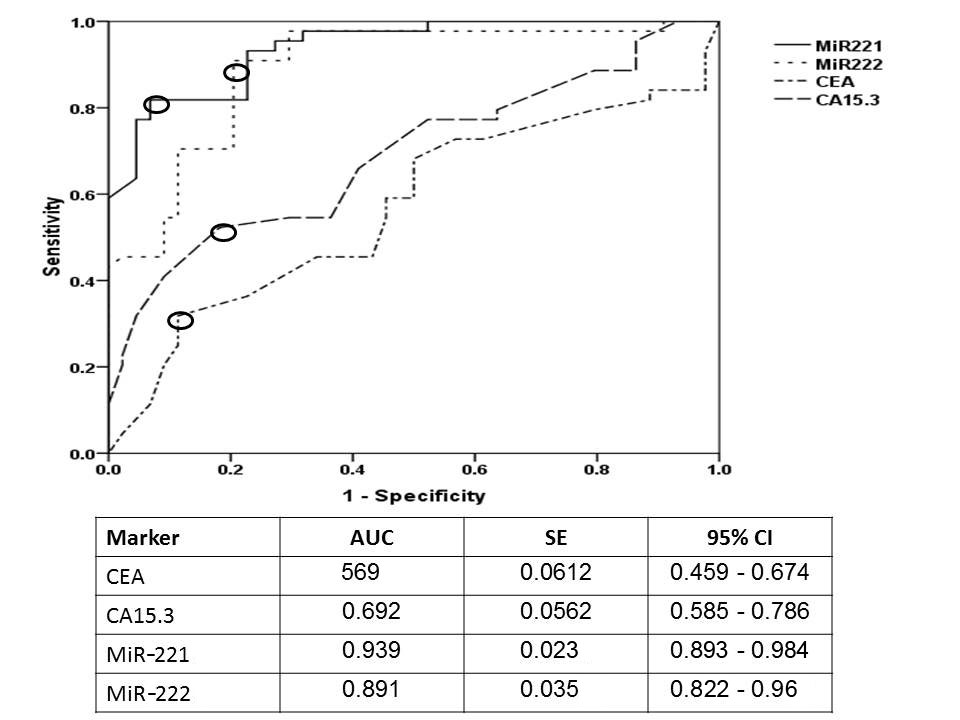 Figure 1. Receiver operating characteristic curve (ROC) for investigated miRNAs and tumor markers. The open circle donates the best cutoff point for investigated markers; for miR-221 was 32.4 fold change with AUC (95%CI, SE) 0.939 (0.893-0.894, 0.023) at P<0.001, for miR-222 was 21.6 fold change with AUC 0.891 (0.822-0.96, 0.035) at P<0.001, for CEA 15.3 ng/ml with AUC 0.569 (0.459 - 0.674, 0.0612) at P=0.018, and for CA15.3 was 21 ng/ml with AUC 0.692 (0.585 - 0.786,0.0562) at P<0.0001.
Figure 1. Receiver operating characteristic curve (ROC) for investigated miRNAs and tumor markers. The open circle donates the best cutoff point for investigated markers; for miR-221 was 32.4 fold change with AUC (95%CI, SE) 0.939 (0.893-0.894, 0.023) at P<0.001, for miR-222 was 21.6 fold change with AUC 0.891 (0.822-0.96, 0.035) at P<0.001, for CEA 15.3 ng/ml with AUC 0.569 (0.459 - 0.674, 0.0612) at P=0.018, and for CA15.3 was 21 ng/ml with AUC 0.692 (0.585 - 0.786,0.0562) at P<0.0001.
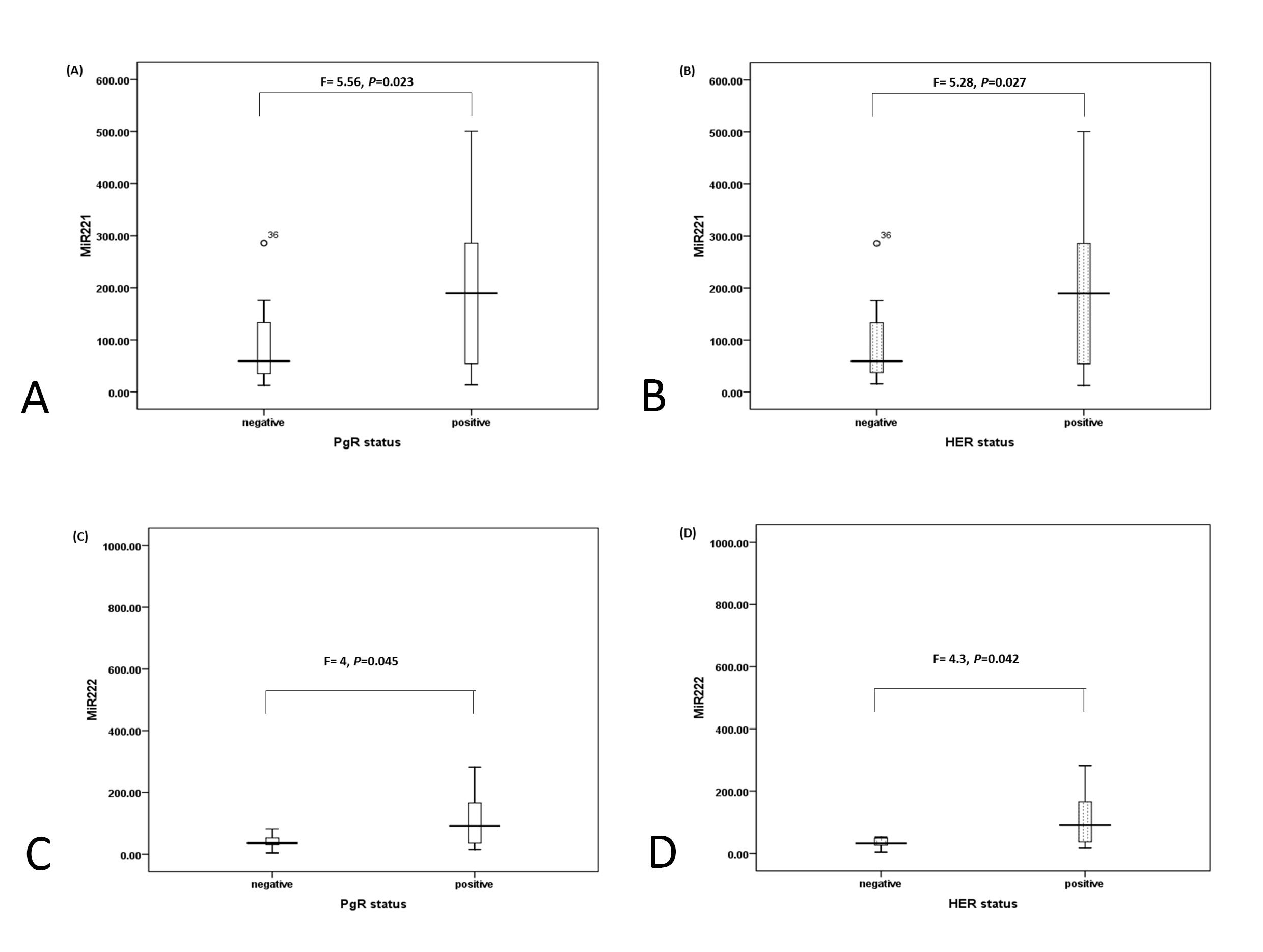 Figure 2. Box-Plot represents the relation between hormonal status and miR-221 and miR - 222. (A) Relation between miR-221 and PgR as mean level± SEM is 95 ± 23 for negative PgR and 191 ± 26.6 for positive PgR. (B) Relation between miR-221 and HER-2/neu as mean level ± SEM is 96 ±23 for negative HER-2 and 190 ± 26.7 for positive HER-2. (C) Relation between miR-222 and PgR as mean level ± SEM is 46.7± 7.7 for negative PgR and 128.3 ± 29.3 for positive PgR. (D) Relation between miR-222 and HER-2/neu as mean level ± SEM is 44.2 ±8 for negative HER-2 and 129 ± 29 for positive HER-2.
Figure 2. Box-Plot represents the relation between hormonal status and miR-221 and miR - 222. (A) Relation between miR-221 and PgR as mean level± SEM is 95 ± 23 for negative PgR and 191 ± 26.6 for positive PgR. (B) Relation between miR-221 and HER-2/neu as mean level ± SEM is 96 ±23 for negative HER-2 and 190 ± 26.7 for positive HER-2. (C) Relation between miR-222 and PgR as mean level ± SEM is 46.7± 7.7 for negative PgR and 128.3 ± 29.3 for positive PgR. (D) Relation between miR-222 and HER-2/neu as mean level ± SEM is 44.2 ±8 for negative HER-2 and 129 ± 29 for positive HER-2.
In the present study, expression level of MiRNA-221and miRNA-222 were detected in serum samples from a total of 88 individuals grouped according to their clinical diagnosis into patients with primary breast cancer, patients with benign breast lesions, and healthy individuals served as control, the results compared to other commonly used protein-based markers (CEA and CA15-3) to assess the role of miRNAs as diagnostic markers for breast cancer. Currently, both MiRNA-221and miRNA-222 were up-regulated in breast cancer group with high median levels (119) for miRNA-221 and (52.8) for miRNA-222 compared to other groups (benign and healthy control), a result in harmony with Kim et al.38 and in line with Swellam et al 10, 13 for miRNA-222. An in vitro study by Falkenberg and his colleagues 39 stated that overexpression of miRNA-221and miRNA-222 induced cell proliferation and invasion. The diagnostic efficacy was detected using ROC curve and revealed high AUC (0.939) and (0.891) for miRNA-221 and miRNA-222, respectively compared to routine tumor markers. These results indicated the oncogenic properties of miRNA-221 and miRNA-222 that may be due to their role as promoting malignant cell proliferation through suppressing cyclin-dependent kinase inhibitors expression regulators of cell cycle progression [40]. The present results in concordance with a study carried using breast tissue samples by Zong and his colleagues [36] whose stated that the expression level of miRNA-221and miRNA-222 increased in breast cancer tissues compared with non-cancerous tissues.
The relation between investigated markers with clinicopathological factors revealed that miRNA-221and miRNA-222 expressions reported statistically significant increase in late stages (III-IV) and high-grade tumors (III). Moreover their expression increased significantly with lymph nodes involvement, which are in concordance of previous studies [39, 41] that emphasize their involvement in breast cancer progression and metastasis, which is one of the critical causes of breast cancer patient’s death. The spread of primary breast tumor cells to the lymph nodes is at the forefront of symbolizing the first signs of metastatic expansion [42] thus, aberrant expression of molecular markers in BC tissues with lymph node metastases may represent early biomarkers of the risk for developing distant metastases.
Overexpression of miRNA-221and miRNA-222 significantly increased in breast cancer patients with IDC compared to non-IDC which indicates that it can be used to discriminate between different pathological types; this may be due to its negative correlation with inhibitor matrix metalloproteinase −3 (TIMP-3) which have anti-invasive [43] and anti-angiogenic features [44].
In breast cancer, miRNA-221and miRNA-222 was more abundant in basal-like tumors than in ER/PR–positive tumors 44 miRNA-221and miRNA-222 directly target ER-alpha gene transcription, which is implicated in breast tumorgenesis, especially more aggressive basal-like breast cancer [46, 47], hence the ER is inhibited upon overexpression of miRNA-221and miRNA-222 and cancer cells grow in an estrogen independent mode. Current study revealed non-significant difference between hormonal ER- status and miRNA-221and miRNA-222, which may postulate that this group of breast cancer patients may not benefit from tamoxifen, a traditional anti-estrogen that competes with ER for ER-α [48].On the other hand this study reported significant difference between PgR and HER-2/neu positive levels with miRNA-221 and miRNA-222, which may suggest their possible role as predictive prognostic marker for this type of cancer.
Currently, significant positive correlation was reported between miRNA-221 and miRNA-222 and this may be due to the fact that they are homologues miRs in human DNA, chromosome Xp11.3 is the miRNA-221and miRNA-222 gene cluster site [49] and hence they both may contribute in breast cancer aggressiveness.
As reported in Table 3, the sensitivities and PPV values for miRNA-221 and miRNA-222 were superior to CEA and CA15.3 for detection of breast cancer as well as for identification of breast cancer patients with early stages and low grades. These finding indicate the prospective role of using miRNA-221 and miRNA-222 as early diagnostic molecular markers in breast cancer.
Upon detection of breast cancer patients, our results emphasize high significant expression of miRNA-221 and miRNA-222 in serum samples of breast patients as compared to routinely used blood-based tumor markers, CEA and CA15.3, these results point out the value of miRNA-221 and miRNA-222 as noninvasive markers for early detection of breast cancer patients especially those at high-risk groups.
This work was supported equally through a grant from Science Technology Development Fund (STDF) through Basic and Applied Research Support Grant Project (BARG) [No.15089], Egypt, and as part from PhD thesis submitted to Zoology Department, Faculty of Science (Girls), Al-Azhar University, Egypt.
The instruments listed in the current study were purchased through a grant from Science Technology Development Fund (STDF) through Capacity Building Grant Project (CBG) [No. 4940]
Funding
This research did not receive any specific financial support from funding agencies in the public, commercial, or not-for-profit sectors.
Author contributions
Study conception and design: by MS, EAA, SAA and MAM; Acquisition of data: EAA and MAM; Analysis and interpretation of data: MS and EAA; Drafting of manuscript: MS, EAA, SAA and MAM; Critical revision: MS, EAA, SAA and MAM.
Competing interests
The authors declare no conflict of interest with the work.
Ethical statement
Ethical approval from the Scientific Medical Ethical Committee (National Research Centre, number#15029) was obtained for the current study.
- Abdel-Salam IM, Ashmawy AM, Hilal AM, Eldahshan OA, Ashour M: Chemical Composition of Aqueous Ethanol Extract of Luffa cylindrical Leaves and Its Effect on Representation of Caspase-8, Caspase-3, and the Proliferation Marker Ki67 in Intrinsic Molecular Subtypes of Breast Cancer in Vitro. Chem Biodivers 2018, 15(8): e1800045.
- Bray F, Ferlay J, Soerjomataram I, Siegel Rl, Torre lA, Jemal A: Global Cancer Statistics 2018: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2018, 68(6): 394-424.
- Assi H, Khoury KE, Dbouk H: Epidemiology and Prognosis of Breast Cancer in Young Women. J Thorac Dis 2013, 5 Suppl 1: S2-8.
- Ibrahim AS, Khaled HM, Mikhail NN, Baraka H, Kamel H: Cancer Incidence in Egypt: Results of the National Population-Based Cancer Registry Program. J Cancer Epidemiol 2014, 2014: 437971.
- Zhang L, Xu Y, Jin X, Wang Z, Wu Y, Zhao D, Chen G, Li D, Wang X, Cao H, et al: A circulating miRNA signature as a diagnostic biomarker for non-invasive early detection of breast cancer. Breast Cancer Res Treat 2015, 154(2): 423-434.
- Hamouda SKM, Abo El-Ezz RH, Wahed ME: Enhancement Accuracy of Breast Tumor Diagnosis in Digital Mammograms. J Biomed Sci 2017, 6(4): 28.
- Patani N, Martin LA, Dowsett M: Biomarkers for the clinical management of breast cancer: international perspective. Int J Cancer 2013, 133 (1): 1-3.
- Ng EKO, Li R, Shin VY, Siu JM, Ma ESK, Kwong A: MicroRNA-143 is downregulated in breast cancer and regulates DNA methyltransferases 3A in breast cancer cells. Tumor Biology 2014, 35(3): 2591-2598.
- Chen YX, Huanga bKJ, Niu KX: Recent advances in signal amplification strategy based on oligonucleotide and nanomaterials for microRNA detection-a review. Biosens Bioelectron 2018, 15(99): 612-624.
- Swellam M, Zahran RFK, Taha HA, El-Khazragy N, Abdel-Malak C: Role of some circulating MiRNAs on breast cancer diagnosis. Arch Physiol Biochem 2018(b), 20: 1-9.
- Zhang DQ, Zhou CK, Jiang XW, Chen J, Shi BK: Increased expression of miR-222 is associated with poor prognosis in bladder cancer. World J Surg Oncol 2014, 12: 241.
- Jurkovicova D, Smolkova B, Sestakova Z, Kalinkova L: Down-regulation of traditional oncomiRs in plasma of breast cancer patients. Oncotarget 2017, 8(44): 77369-77384.
- Swellam M, El Magdoub HM, Hassan, NM, Hefny, MM, Sobeih ME: Potential diagnostic role of circulating MiRNAs in breast cancer: Implications on clinicopathological characters. Clin Biochem 2018(a), 56: 47-54.
- Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G: MiR221/222in cancer: their role in tumor progression and response to therapy. Curr Mol Med 2012, 12(1): 27-33.
- Howe EN, Cochrane DR, Richer JK: The miR-200 and miR-221/222microRNA families: opposing effects on epithelial identity. J Mammary Gland Biol Neoplasia 2012, 17(1): 65-77.
- Amini S, Abak A, Estiar MA, Montazeri V, Abhari A, Sakhinia E: Expression Analysis of MicroRNA-222 in Breast Cancer. Clin Lab 2018, 64(4): 491-496.
- Zhou Y, Sheng B, Xia Q, Guan X, Zhang Y: Association of long non-coding RNA H19 and microRNA-21 expression with the biological features and prognosis of non-small cell lung cancer. Cancer Gene Ther 2017, 24(8): 317-324.
- le Sage C, Nagel R, Egan DA, Schrier M, Mesman E, Mangiola A, Anile C, Maira G, Mercatelli N, Ciafrè SA, et al: Regulation of the p27(Kip1) tumor suppressor by miR-221 and miR-222 promotes cancer cell proliferation. EMBO J 2007, 26(15): 3699-3708.
- Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, Taccioli C, Pichiorri F, Alder H, Secchiero P, et al: miR221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell 2009, 16(6): 498-509.
- Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H, et al: MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst 2010, 102(10): 706-721.
- Li B, Lu Y, Wang H, Han X, Mao J, Li J,Yu L,Wang B, Fan S, Yu X, Song B: miR-221/222 enhance the tumorigenicity of human breast cancer stem cells via modulation of PTEN/Akt pathway. Biomed Pharmacother 2016, 79: 93-101.
- Greene FL, Sobin LH: The staging of cancer: a retrospective and prospective appraisal. CA Cancer J Clin 2008, 58(3): 180-190.
- Robbins P, Pinder S, de Klerk N, Dawkins H, Harvey J, Sterrett G, Ellis I, Elston C: Histological grading of breast carcinomas: a study of interobserver agreement. Hum Pathol 1995, 26(8): 873-879.
- Hsu SM, Raine L, Fanger H: A comparative study of the peroxidase-anti-peroxidase method and an avidin-biotin complex method for studying polypeptide hormones with radioimmunoassay antibodies. Am J Clin Pathol 1981, 75(5): 734-738.
- Kim SH, Seo B, Lee J: Correlation of ultra-sound findings with histology, tumor grade, and biological markers in breast cancer. Acta Oncol 2008, 47(8): 1531-1538.
- Livak KJ, Schmittgen TD: Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta DeltaC (T) method. Methods 2001, 25(4): 402-408.
- Zweig MH, Campbell G: Receiver-operating characteristic (ROC) plots: a fundamental evaluation tool in clinical medicine. Clin Chem 1993, 39(4): 561-577.
- Hamam R, Hamam D, Alsaleh KA, Kassem M, Zaher W, Alfayez M, Aldahmash A, Alajez NM: Circulating microRNAs in breast cancer: novel diagnostic and prognostic biomarkers. Cell Death Dis 2017, 8(9): e3045.
- He Y, Deng F, Yang S, Wang D, Chen X, Zhong S, Zhao J, Tang J: Exosomal microRNA: a novel biomarker for breast cancer. Biomark Med 2018, 12(2): 177-188.
- El-Khazragy N, Noshi MA, Abdel-Malak C, Zahran RF, Swellam M: MiRNA-155 and miRNA-181a as prognostic biomarkers for pediatric acute lymphoblastic leukemia. J Cell Biochem 2019, 120(4): 6315-6321.
- Wurz K, Garcia RL, Goff BA, Mitchell PS, Lee JH, Tewari M, Swisher EM: MiR-221 and MiR-222 alterations in sporadic ovarian carcinoma: Relationship to CDKN1B, CDKNIC and overall survival. Genes Chromosomes Cancer 2010, 49(7): 577-584.
- Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, Mazzaferro V, Lowe SW, Croce CM, Dejean A: MiR-221 over expression contributes to liver tumorigenesis. Proc Natl Acad Sci USA 2010, 107(1): 264-269.
- Zhang J, Han L, Ge Y, Zhou X, Zhang A, Zhang C, Zhong Y, You Y, Pu P, Kang C: MiR-221/222 promote malignant progression of glioma through activation of the Akt pathway. Int J Oncol 2010, 36(4): 913-20.
- Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, Burow ME, Ivan M, Croce CM, Nephew KP: MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene 2011, 30(9): 1082-1097.
- Chun-Zhi Z, Lei H, An-Ling Z, Yan-Chao F, Xiao Y, Guang-Xiu W, Zhi-Fan J, Pei-Yu P, Qing-Yu Z, Chun-Sheng K: MicroRNA-221 and microRNA-222 regulate gastric carcinoma cell proliferation and radioresistance by targeting PTEN. BMC Cancer 2010, 10: 367.
- Zong Y, Zhang Y, Sun X, Xu T, Cheng X, Qin Y: MiR-221/222 promote tumor growth and suppress apoptosis by targeting lncRNA GAS5 in breast cancer. Biosci Rep 2019, 39(1): pii: BSR20181859.
- Roscigno G, Quintavalle C, Donnarumma E, Puoti I, Diaz-Lagares A, Iaboni M, Fiore D, Russo V, Todaro M, Romano G, et al: MiR-221 promotes stemness of breast cancer cells by targeting DNMT3b. Oncotarget 2016, 7(1): 580-592.
- Kim J, Oh S, Park S, Ahn S, Choi Y, Kim G, Kim SI, Lee H: Circulating miR-221 and miR-222 as Potential Biomarkers for Screening of Breast Cancer. Biomed Sci Letters 2019, 25(2): 185-189.
- Falkenberg N, Anastasov N, Rappl K, Braselmann H, Auer G, Walch A, Huber M, Höfig I, Schmitt M, Höfler H, et al: MiR-221/-222 differentiates prognostic groups in advanced breast cancers and influence cell invasion. Br J Cancer 2013, 109(10): 2714-2723.
- Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, Farace MG: MiR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem 2007, 282 (32): 23716-23724.
- Chernyy V, Pustylnyak V, Kozlov V, Gulyaeva L: Increased expression of miR-155 and miR-222 is associated with lymph node positive status. J Cancer 2018, 9(1): 135-140.
- Eccles S, Paon L, Sleeman J: Lymphatic metastasis in breast cancer: importance and new insights into cellular and molecular mechanisms. Clin Exp Metastasis 2007, 24(8): 619-636.
- Celebiler Cavusoglu A, Kilic Y, Saydam S, Canda T, Başkan Z, Sevinc AI, Sakizli M: Predicting invasive phenotype with CDH1, CDH13, CD44, and TIMP3 gene expression in primary breast cancer. Cancer Sci 2009, 100(12): 2341-2345.
- Qi JH, Ebrahem Q, Moore N, Murphy G, Claesson-Welsh L, Bond M, Baker A, Anand-Apte B: A novel function for tissue inhibitor of metallo-proteinases-3 (TIMP3): inhibition of angiogenesis by blockage of VEGF binding to VEGF receptor-2. Nat Med 2003, 9(4): 407-415.
- Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O'Brien C, Spoerke J, Jhunjhunwala S, Boyd Z, Januario T, et al: TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal 2011, 4(177): ra41.
- Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, Coppola D, Cheng JQ: MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem 2008, 283(45): 31079-31086.
- Kumar R, Jin X, Zhen Y, Hu P: MicroRNA regulates estrogen receptor alpha in breast Cancer metastasis. J Cancer Res Therap Oncol 2014, 2: 1-6.
- Jordan VC, Obiorah I, Fan P, Kim HR, Ariazi E, Cunliffe H, Brauch H: The St. Gallen prizelecture 2011: evolution of long-term adjuvant anti-hormone therapy: consequences and opportunities. Breast 2011, 20 Suppl. 3: S1-S11.
- Abak A, Amini S, Sakhinia E, Abhari A: MicroRNA-221: biogenesis, function and signatures in human cancers. Eur Rev Med Pharmacol Sci 2018, 22(10): 3094-3117.
Asia-Pacific Journal of Oncology
print ISSN: 2708-7980, online ISSN: 2708-7999
 Copyright © Asia Pac J Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Asia Pac J Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript