Research Article | Open Access
Implication of expression of MMR proteins and clinicopathological characteristics in gastric cancer
Renu Verma1, Puja Sakhuja2, Ritu Srivastava2, Prakash Chand Sharma11University School of Biotechnology, Guru Gobind Singh Indraprastha University, New Delhi, India.
2Department of Pathology, Govind Ballabh Pant Hospital, New Delhi, India.
Correspondence: Prakash C. Sharma ( University School of Biotechnology, Guru Gobind Singh Indraprastha University Dwarka, Sec 16C, New Delhi 110078, India ; E-mail: prof.pcsharma@gmail.com).
Asia-Pacific Journal of Oncology 2020, 1: 61-67. https://doi.org/10.32948/ajo.2020.12.30
Received: 11 Dec 2020 | Accepted: 29 Dec 2020 | Published online: 30 Dec 2020
Introduction Microsatellite instability (MSI), referred to as variations at microsatellite loci, at mismatch repair (MMR) genes leads to the formation of an aberrant MMR system that fails to rectify errors occurring during DNA replication. MMR deficiency can be assessed by immunohistochemical analysis of the expression of mismatch repair proteins in the target tissues.
Methods We investigated the expression of four key MMR proteins (MLH1, MSH2, MSH6, and PMS2) in formalin-fixed paraffin-embedded (FFPE) tumor and normal tissues obtained from thirty gastric cancer (GC) patients. The association of clinicopathological features with MMR status was also analyzed.
Results A total of 12 (40%) GC patients exhibited loss of expression of MMR proteins, including loss of MLH1 and PMS2 in 3 cases and loss of MSH2 and MSH6 in 4 cases. Univariate analysis showed an association of loss of MMR protein expression with moderately differentiated GC. However, there was no statistically significant association between loss of MMR protein expression with gender, tumor location, depth of invasion, lymph node metastasis, WHO classification, lympho-vascular invasion, and infection with H. pylori.
Conclusion Our results implicate the role of mismatch repair proteins in gastric tumorigenesis. The MMR protein status is an important aspect of tumorigenesis and can be prescribed for the screening of GC.
Key words Mismatch repair proteins, Immunohistochemistry, Gastric cancer, H. pylori
Microsatellites, popularly known as Simple Sequence Repeats (SSRs), are short iterations of 1-6 base long units. Instability at microsatellite locus is caused by loss of mismatch repair machinery, replication errors, or polymerase slippage. Over the last two decades, Microsatellite Instability (MSI) has been investigated widely for its implication in different cancers. In 1997, a panel of five microsatellite markers consisting of two mononucleotide repeats (BAT25 and BAT26) and three dinucleotide repeats (D2S123, D5S346, and D17S250) was recommended by the National Cancer Institute for the evaluation of Lynch syndrome [6]. MSI accounts for 22% of the tumors and has been classified as one of the subgroups of gastric cancers by TCGA [7]. Mismatch repair genes rectify mistakes that occur in the genome by DNA exonuclease proof-reading ability. Mutations in these genes hamper the correction work in the genome, which leads to the increase in mutation rate and contributes towards oncogenesis. Instability in these genes affects cancer initiation and progression by changing their expression [8]. The major proteins involved in the DNA mismatch repair mechanism are MLH1, MSH2, MSH6, and PMS2 [9, 10]. MSI has earlier been reported in various cancers like colorectal, endometrial, ovarian, and gastric cancer [11-13].
Loss of MMR protein expression has been recorded in up to 30% of GC cases [14-18]. In the present study, we have reported the expression of four mismatch repair proteins, MLH1, MSH2, MSH6, and PMS2 in GC using an immunohistochemical staining procedure in Indian population along with the association of clinical parameters of GC patients with loss of expression of mismatch repair proteins.
Tumor and adjacent normal tissue samples from thirty GC patients were included in the study. All tissue samples were pathologically examined, and the diagnosis of gastric cancer was confirmed in the department of pathology, Govind Ballabh Pant Hospital, New Delhi. Written consent was taken from all the participants. The clinical parameters of all patients were recorded.
Immunohistochemistry
Thirty paraffin-embedded sections (2 µm) of histologically confirmed gastric cancer along with normal tissues were obtained on poly L-lysine coated blocks. Briefly, the tissue sections were deparaffinized at 60ºC for 20 min and rehydrated twice with xylene and thrice with acetone for 5 min each. Antigen retrieval was carried out with Tris-EDTA buffer at 98ºC for 20 min, followed by 3X TBS wash for 2 min each. The tissue sections were incubated with H2O2 for 20 min followed by 3X TBS wash for 2 min each. Later, protein block was added, and the tissue slides were incubated overnight with 1:100 diluted mouse monoclonal anti-MLH1antibody (Thermo Scientific, USA), 1:10 diluted mouse monoclonal anti-MSH2 antibody (Thermo Scientific, USA), ready to use rabbit monoclonal anti-MSH6 antibody (DAKO, USA) and ready to use rabbit monoclonal anti-PMS2 antibody (DAKO, USA) of human origin at 4ºC.
The next day, the slides were washed 3X with TBS saline for 2-3 min each and coated with primary antibody amplifier Quanto (Thermo Scientific, USA) for 15 min. Again, 3X wash was given with TBS for 2 min each. Then, a secondary antibody (HRP polymer Quanto, Thermo Scientific, USA) was added for 1 h, followed by 3X washing with TBS for 2 min. The color was developed using DAB as a chromogen for 10 min. Finally, the slides were washed with distilled water and mounted with DPX. GC tissue sections not treated with anti-MLH1, anti-MSH2, anti-MSH6, and anti-PMS2 were used as negative controls. MLH1, MSH2, MSH6, and PMS2 protein expression were counted as positive if epithelial cells showed immunopositivity in the nucleus. The slides were independently screened by two pathologists.
Statistical analysis
Data recorded were statistically analyzed by using the Statistical Program for Social Sciences (SPSS) version 25.0 (SPSS Inc., Chicago, IL, USA). The association between clinicopathological parameters of GC patients and MMR protein expression was analyzed by student t-test or χ2 test. A p ≤ 0.05 was considered a criterion for statistical significance.
Out of 30, in total, 12 (40%) GC patients exhibited the loss of expression of one or more MMR proteins investigated in the present study. Representative immunohistochemical sections of all the four proteins with loss and intact expression are shown in Figure 1 (MLH1 and PMS2) & Figure 2 (MSH2 and MSH6). Briefly, 16.7% loss of expression was observed in MLH1, MSH2, and PMS2, while MSH6 showed a loss in 30% cases. All the observed combinations of loss of expression are given in Table 1.
We analyzed clinicopathological features associated with MSI tumor phenotypes. There was no difference in MMR-positive and MMR-negative GC patients with respect to age (59.28 ± 12.65, 62.58 ± 9.5, p= 0.067). All MMR negative gastric cancer patients were 60 years old and above except two patients. Only one female patient was present in the group who was MMR negative. Out of twelve, nine MMR negative tumors were located in the cardia, fundus, and body. The depth of invasion, according to TNM classification, was T1 for one case and T2 for another, seven cases of T3, and three cases of T4 in MSI tumors. Lymph node metastasis was seen in 75% of MMR negative tumors. There was no metastatic tumor in the group studied for MMR protein expression. One tumor was well-differentiated; nine tumors were moderately differentiated, while two were poorly differentiated. Haematoxylin and eosin-stained sections of well-differentiated, moderately differentiated, and poorly differentiated tumors are represented in Figure 3. Lymphocytic invasion was present in nine MMR negative tumors. One case of papillary, 4 cases of tubular, 4 cases of poorly cohesive adenocarcinomas were categorized histologically by WHO classification. One MMR negative case showed positive H. pylori infection (Figure 4). Univariate analysis for association of clinicopathological features and MMR protein expression is summarized in Table 2.
Loss of MMR expression exhibited predominant association with moderately differentiated type in the histology of GC. However, gender, tumor location, depth of invasion, lymph node metastasis, WHO classification, lympho-vascular invasion, and H. pylori infection were not associated with negative MMR protein immunohistochemical expression.
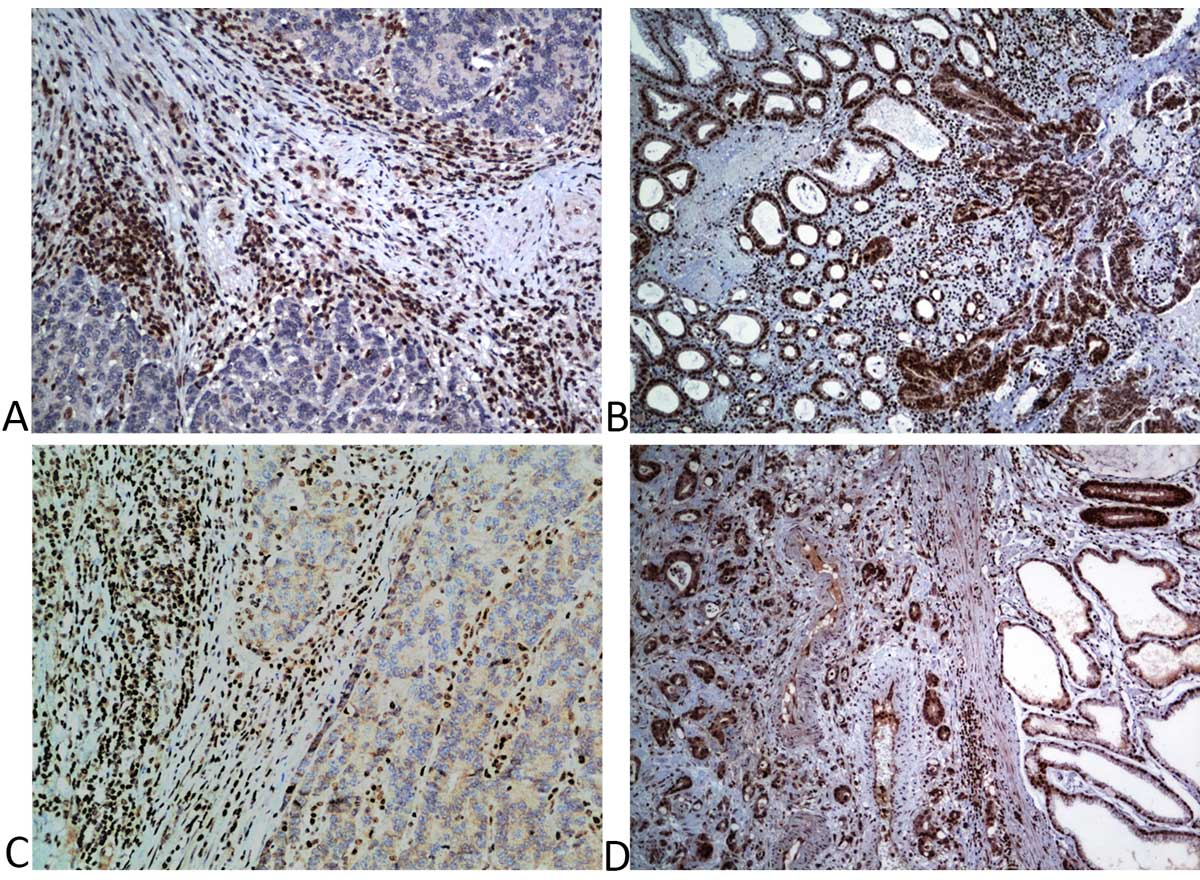 Figure 1. Immunohistochemical staining for the detection of mismatch repair (MMR) proteins in gastric cancer (GC) (a) Loss of nuclear expression of MLH1 (DAB; x20); (b) Intact nuclear expression of MLH1 (DAB; x10); (c) Loss of nuclear expression of PMS2 with adjacent mucosa as control (DAB; x40); (d) Intact nuclear expression of PMS2 (DAB; x10).
Figure 1. Immunohistochemical staining for the detection of mismatch repair (MMR) proteins in gastric cancer (GC) (a) Loss of nuclear expression of MLH1 (DAB; x20); (b) Intact nuclear expression of MLH1 (DAB; x10); (c) Loss of nuclear expression of PMS2 with adjacent mucosa as control (DAB; x40); (d) Intact nuclear expression of PMS2 (DAB; x10).
 Figure 2. Immunohistochemical staining for the detection of mismatch repair proteins (a) Loss of nuclear expression of MSH2 (DAB; x20); (b) Intact nuclear expression of MSH2 (DAB; x10); (c) Loss of nuclear expression of MSH6 with adjacent normal mucosa (DAB; x20); (d) Intact nuclear expression of MSH6 (DAB; x20).
Figure 2. Immunohistochemical staining for the detection of mismatch repair proteins (a) Loss of nuclear expression of MSH2 (DAB; x20); (b) Intact nuclear expression of MSH2 (DAB; x10); (c) Loss of nuclear expression of MSH6 with adjacent normal mucosa (DAB; x20); (d) Intact nuclear expression of MSH6 (DAB; x20).
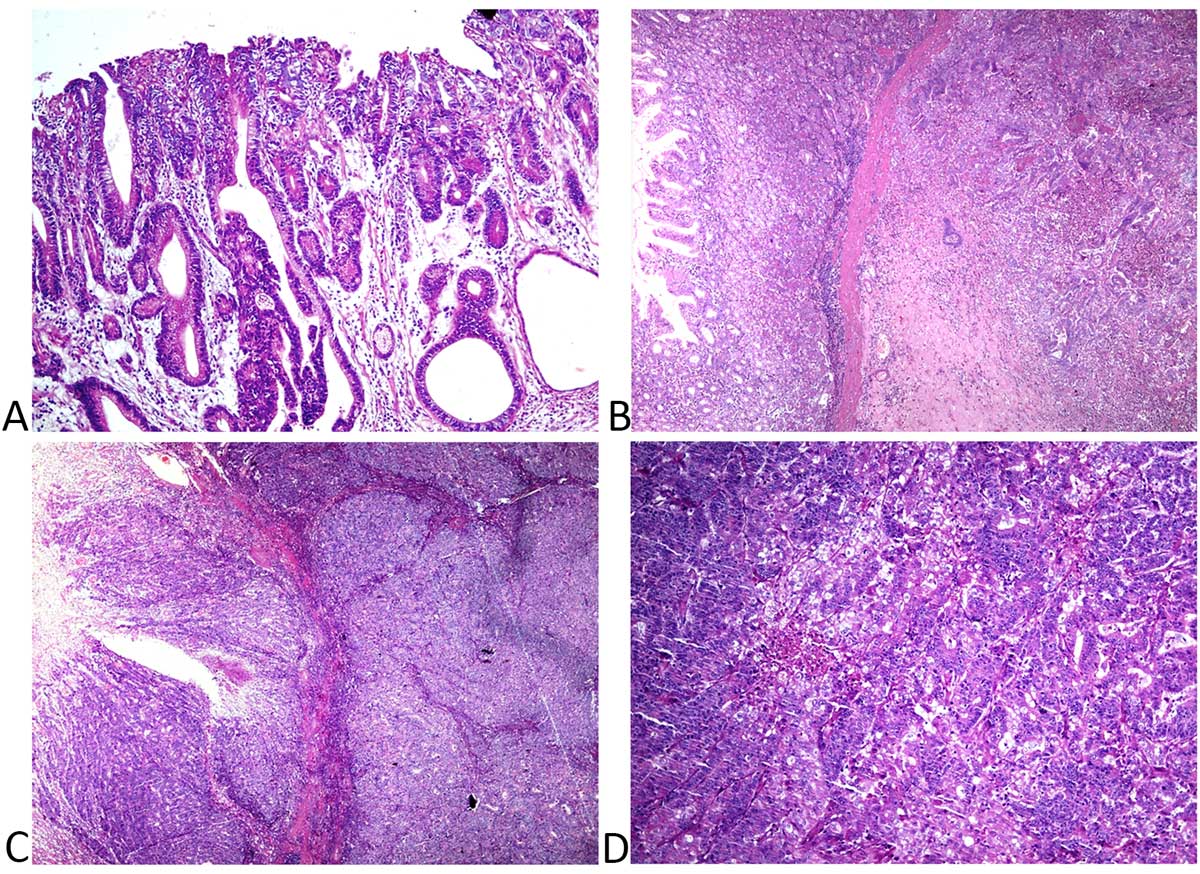 Figure 3. Haematoxylin and Eosin (H&E) stained photomicrograph showing histology of tumor tissues (a) well-differentiated adenocarcinoma of stomach showing the presence of tumor-infiltrating lymphocytes (Magnification x400) (b) H&E stained photomicrograph showing moderately differentiated adenocarcinoma of stomach showing the presence of tumor-infiltrating lymphocytes (Magnification x200) (c, d) H&E stained photomicrograph showing poorly cohesive adenocarcinoma of stomach and presence of tumor-infiltrating lymphocytes (Magnification x200 and x400, respectively).
Figure 3. Haematoxylin and Eosin (H&E) stained photomicrograph showing histology of tumor tissues (a) well-differentiated adenocarcinoma of stomach showing the presence of tumor-infiltrating lymphocytes (Magnification x400) (b) H&E stained photomicrograph showing moderately differentiated adenocarcinoma of stomach showing the presence of tumor-infiltrating lymphocytes (Magnification x200) (c, d) H&E stained photomicrograph showing poorly cohesive adenocarcinoma of stomach and presence of tumor-infiltrating lymphocytes (Magnification x200 and x400, respectively).
 Figure 4. Giemsa staining of gastric tumor tissue section showing infection with Helicobacter pylori.
Figure 4. Giemsa staining of gastric tumor tissue section showing infection with Helicobacter pylori.
|
Table 1. Expression of MMR proteins (MLH1, MSH2, MSH6 and PMS2) in 30 GC patients. |
|
|
MSI status |
No. of patient (%) |
|
MSS |
18 (60.0) |
|
MSI |
12 (40.0) |
|
Loss of MLH1 |
5 (16.7) |
|
Loss of MSH2 |
5 (16.7) |
|
Loss of MSH6 |
9 (30.0) |
|
Loss of PMS2 |
5 (16.7) |
|
Loss of MLH1 and MSH2 |
1 (3.3) |
|
Loss of MLH1 and MSH6 |
3 (10.0) |
|
Loss of MLH1 and PMS2 |
3 (10.0) |
|
Loss of MSH2 and MSH6 |
4 (13.3) |
|
Loss of MSH2 and PMS2 |
1 (3.3) |
|
Loss of MSH6 and PMS2 |
2 (6.7) |
|
Loss of MLH, MSH2, MSH6 |
1 (3.3) |
|
Loss of MSH2, MSH6, PMS2 |
1 (3.3) |
|
Loss of MSH6, PMS2, MLH1 |
1 (3.3) |
|
Loss of PMS2, MLH1, MSH2 |
0 (0) |
|
MSS: Microsatellite stable; MSI: Microsatellite instability; MLH1: Human Mutl homolog 1; MSH2: Human MutS homolog 2; MSH6 : Human MutS homolog 6; PMS2: PMS1 homolog 2; GC: gastric cancer; MMR: mismatch repair. |
|
|
Table 2. Univariate analysis of clinicopathological characteristics and MMR protein expression in GC. |
||||
|
Variable
|
Total
|
MMR positive |
MMR negative |
p-value |
|
Age |
|
|
|
0.067 |
|
<60 |
11 |
9 (81.8) |
2 (18.2) |
|
|
≥60 |
19 |
9 (47.4) |
10 (52.6) |
|
|
|
|
|
|
|
|
Gender |
|
|
|
0.227 |
|
Male |
29 |
18 (62) |
11 (38) |
|
|
Female |
1 |
0 (0) |
1 (100) |
|
|
|
|
|
|
|
|
Tumor Location |
|
|
|
0.105 |
|
Upper and middle |
17 |
8 (47.1) |
9 (52.9) |
|
|
Lower |
13 |
10 (76.9) |
3 (23.1) |
|
|
|
|
|
|
|
|
Depth of invasion |
|
|
|
0.775 |
|
T1 |
2 |
1 (50) |
1 (50) |
|
|
T2, T3, T4 |
28 |
17 (60.7) |
11 (39.3) |
|
|
|
|
|
|
|
|
Lymph node metastasis |
|
|
|
0.64 |
|
N0 |
9 |
6 (66.7) |
3(33.3) |
|
|
N1, N2, N3 |
21 |
12 (57.2) |
9(42.8) |
|
|
|
|
|
|
|
|
Histology |
|
|
|
0.002 |
|
Well and Moderately differentiated |
15 |
5 (33.3) |
10 (66.7) |
|
|
Poorly differentiated |
15 |
13 (86.7) |
2 (13.3) |
|
|
|
|
|
|
|
|
WHO Classification |
|
|
|
|
|
Papillary + Tubular |
7 |
2 (28.6) |
5 (71.4) |
0.117 |
|
Poorly cohesive |
16 |
12 (75) |
4 (25) |
|
|
Others |
7 |
4 (57.2) |
3 (42.8) |
|
|
|
|
|
|
|
|
Lymphovascular invasion |
|
|
|
0.295 |
|
Present |
19 |
10 (52.6) |
9 (47.4) |
|
|
Absent |
11 |
8 (72.7) |
3 (27.3) |
|
|
|
|
|
|
|
|
H. pylori infection |
|
|
|
0.334 |
|
Present |
5 |
4 (80) |
1 (20) |
|
|
Absent |
25 |
14 (56) |
11 (44) |
|
The work presented here explored the prevalence of loss of MMR proteins in gastric cancer. The percentage (40%) of patients who revealed the loss of one or more MMR proteins is relatively high as compared to other studies demonstrating varying degree (5.6%-30%) of defective MMR expression all over the world in GC [15, 16, 18, 22]. This observation suggests demographic and genetic factors prevailing in human populations contribute towards these variations. We have analyzed loss of expression in 5 cases (16.7%) in MLH1, MSH2, and PMS2 and 9 cases (30%) in MSH6. PMS2 forms a heterodimer with MLH1 only while MLH1 binds with other MMR proteins. Therefore, loss of MLH1 expression leads to loss of PMS2 expression. Similarly, MSH2 and MSH6 form a heterodimer, and loss of MSH2 expression leads to loss of MSH6 expression. Out of all the cases of MMR protein loss, 25% showed loss of MLH1 and PMS2; 33.33% showed loss of MSH2 and MSH6; 16.7% showed isolated loss of PMS2, and 8.3% showed loss of MSH6. Our study contradicted the observation found in most of the related studies that loss of MLH1-PMS2 is prevalent over MSH2-MSH6 loss [18, 23, 24]. Also, previous studies have suggested the predominance in the loss of expression of MLH1 protein over MSH2 protein [15, 17, 25]. However, we observed comparable cases of loss of both MLH1 and MSH2 protein. The loss of MSH6 was found to be present in 75% of MMR negative cases, indicating the significance of the protein among MMR negative GC. One of our earlier reports on gastric cancer also suggested MSH6 gene as an important target for microsatellite instability. This study reinforces the role of MSH6 in the occurrence of gastric cancer [26]. Correlation between different clinical features and loss of MMR protein expression in GC has been analyzed. In gastric cancer, various clinicopathological features, including old age, gender, histological type, intestinal type, advanced lymph node stage, tubular and papillary WHO classification, have been found associated with MMR protein deficiency [17-20]. In our study, we found an association of MMR loss with the moderately differentiated type of histological tissue, which was concordant to another study in GC [20]. Univariate analysis showed that there was no statistically significant association with other selected clinical features in this study.
Out of the twelve cases of negative MMR, ten were of age 60 years or older; eleven were males; nine were located in the upper and middle region of the stomach. Depth of invasion and lymph nodes were on the higher stage in the majority of cases with MMR negative expression. Lymphovascular invasion was seen in 75% of the cases. Only one out of the 5 cases with H. pylori was found to be MMR negative which is different from the findings stating that Helicobacter pylori modulate the MMR expression in ~88% of the GC patients [27-29]. This decline in the outcome may be due to the small population size.
To the best of our knowledge, this is the first study reporting expression of MMR proteins in GC in the Indian population. Our results suggest that expression of MMR plays a crucial role in gastric tumorigenesis as the incidence of loss was quite high in the study, which emphasizes the screening of these proteins to be used as a routine procedure. The loss of expression of MSH6 has been found in three-fourth of patients with MMR negative status in the population studied. The moderately differentiated tumor tissues could be used for better prognosis. This study targeted a small population of GC patients emphasizing the role of screening MMR proteins in India, which needs to be explored in large population size to envisage the prognosis. Our results indicate that MMR expression plays a crucial role in the tumorigenesis of patients diagnosed with gastric cancer.
Ethical clearance was obtained from Maulana Azad Medical College, New Delhi.
Funding
The analysis and interpretation of the manuscript was supported by "CAMS Innovation Fund for Medical Sciences (CIFMS)" and "Platform Improvement of Clinical Trial Capability" (2020-I2M-2-007).
Author contributions
RV designed the study and carried out the sample procurement, immunohistochemical analysis, and prepared the first draft of the manuscript. PS and RS participated in the screening analysis and microphotographs. PCS coordinated the study and finalized the manuscript. All authors read and approved the final manuscript.
Competing interests
All authors disclose no competing interests.
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A: Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018, 68(6): 394-424.
- De Vita F, Di Martino N, Fabozzi A, Laterza MM, Ventriglia J, Savastano B, Petrillo A, Gambardella V, Sforza V, Marano L, et al: Clinical management of advanced gastric cancer: the role of new molecular drugs. World J Gastroenterol 2014, 20(40): 14537-14558.
- Dikshit R, Gupta PC, Ramasundarahettige C, Gajalakshmi V, Aleksandrowicz L, Badwe R, Kumar R, Roy S, Suraweera W, Bray F, et al: Cancer mortality in India: a nationally representative survey. Lancet 2012, 379(9828): 1807-1816.
- Kalyani R, Das S, Kumar ML: Pattern of cancer in adolescent and young adults--a ten year study in India. Asian Pac J Cancer Prev 2010, 11(3): 655-659.
- Cheng XJ, Lin JC, Tu SP: Etiology and Prevention of Gastric Cancer. Gastrointest Tumors 2016, 3(1): 25-36.
- Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al: A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res 1998, 58(22): 5248-5257.
- Comprehensive molecular characterization of gastric adenocarcinoma. Nature 2014, 513(7517): 202-209.
- Ohmiya N, Matsumoto S, Yamamoto H, Baranovskaya S, Malkhosyan SR, Perucho M: Germline and somatic mutations in hMSH6 and hMSH3 in gastrointestinal cancers of the microsatellite mutator phenotype. Gene 2001, 272(1-2): 301-313.
- Shin JS, Tut TG, Yang T, Lee CS: Radiotherapy response in microsatellite instability related rectal cancer. Korean J Pathol 2013, 47(1): 1-8.
- Arends MJ: Pathways of colorectal carcinogenesis. Appl Immunohistochem Mol Morphol 2013, 21(2): 97-102.
- Risinger JI, Berchuck A, Kohler MF, Watson P, Lynch HT, Boyd J: Genetic instability of microsatellites in endometrial carcinoma. Cancer Res 1993, 53(21): 5100-5103.
- Strickler JG, Zheng J, Shu Q, Burgart LJ, Alberts SR, Shibata D: p53 mutations and microsatellite instability in sporadic gastric cancer: when guardians fail. Cancer Res 1994, 54(17): 4750-4755.
- Murphy MA, Wentzensen N: Frequency of mismatch repair deficiency in ovarian cancer: a systematic review This article is a US Government work and, as such, is in the public domain of the United States of America. Int J Cancer 2011, 129(8): 1914-1922.
- Kim JY, Shin NR, Kim A, Lee HJ, Park WY, Kim JY, Lee CH, Huh GY, Park DY: Microsatellite instability status in gastric cancer: a reappraisal of its clinical significance and relationship with mucin phenotypes. Korean J Pathol 2013, 47(1): 28-35.
- Ribeiro U Jr, Jorge UM, Safatle-Ribeiro AV, Yagi OK, Scapulatempo C, Perez RO, Corbett CE, Alves VA, Zilberstein B, Gama-Rodrigues J: Clinicopathologic and immunohistochemistry characterization of synchronous multiple primary gastric adenocarcinoma. J Gastrointest Surg 2007, 11(3): 233-239.
- Warneke VS, Behrens HM, Haag J, Balschun K, Böger C, Becker T, Ebert MP, Lordick F, Röcken C: Prognostic and putative predictive biomarkers of gastric cancer for personalized medicine. Diagn Mol Pathol 2013, 22(3): 127-137.
- Lee HJ, Jang YJ, Lee EJ, Kim JH, Park SS, Park SH, Kim CS, Mok YJ: The significance of mismatch repair genes in gastric cancer. J Cancer Res Ther 2013, 9(1): 80-83.
- Karpińska-Kaczmarczyk K, Lewandowska M, Ławniczak M, Białek A, Urasińska E: Expression of Mismatch Repair Proteins in Early and Advanced Gastric Cancer in Poland. Med Sci Monit 2016, 22: 2886-2892.
- Mathiak M, Warneke VS, Behrens HM, Haag J, Böger C, Krüger S, Röcken C: Clinicopathologic Characteristics of Microsatellite Instable Gastric Carcinomas Revisited: Urgent Need for Standardization. Appl Immunohistochem Mol Morphol 2017, 25(1): 12-24.
- Bae YS, Kim H, Noh SH, Kim H: Usefulness of Immunohistochemistry for Microsatellite Instability Screening in Gastric Cancer. Gut Liver 2015, 9(5): 629-635.
- Seo HM, Chang YS, Joo SH, Kim YW, Park YK, Hong SW, Lee SH: Clinicopathologic characteristics and outcomes of gastric cancers with the MSI-H phenotype. J Surg Oncol 2009, 99(3): 143-147.
- Hayashi T, Arai M, Ueno M, Kinoshita H, Tada Y, Koizumi K, Miki Y, Yamaguchi T, Kato Y, Utsunomiya J, et al: Frequency of immunohistochemical loss of mismatch repair protein in double primary cancers of the colorectum and stomach in Japan. Dis Colon Rectum 2006, 49(10 Suppl): S23-29.
- Kaur G, Masoud A, Raihan N, Radzi M, Khamizar W, Kam LS: Mismatch repair genes expression defects & association with clinicopathological characteristics in colorectal carcinoma. Indian J Med Res 2011, 134(2): 186-192.
- Yuan L, Chi Y, Chen W, Chen X, Wei P, Sheng W, Zhou X, Shi D: Immunohistochemistry and microsatellite instability analysis in molecular subtyping of colorectal carcinoma based on mismatch repair competency. Int J Clin Exp Med 2015, 8(11): 20988-21000.
- Choi JS, Kim MA, Lee HE, Lee HS, Kim WH: Mucinous gastric carcinomas: clinicopathologic and molecular analyses. Cancer 2009, 115(15): 3581-3590.
- Verma R, Agarwal AK, Sakhuja P, Sharma PC: Microsatellite instability in mismatch repair and tumor suppressor genes and their expression profiling provide important targets for the development of biomarkers in gastric cancer. Gene 2019, 710: 48-58.
- Santos JC, Brianti MT, Almeida VR, Ortega MM, Fischer W, Haas R, Matheu A, Ribeiro ML: Helicobacter pylori infection modulates the expression of miRNAs associated with DNA mismatch repair pathway. Mol Carcinog 2017, 56(4): 1372-1379.
- Park DI, Park SH, Kim SH, Kim JW, Cho YK, Kim HJ, Sohn CI, Jeon WK, Kim BI, Cho EY, et al: Effect of Helicobacter pylori infection on the expression of DNA mismatch repair protein. Helicobacter 2005, 10(3): 179-184.
- Kim JJ, Tao H, Carloni E, Leung WK, Graham DY, Sepulveda AR: Helicobacter pylori impairs DNA mismatch repair in gastric epithelial cells. Gastroenterology 2002, 123(2): 542-553.
Asia-Pacific Journal of Oncology
print ISSN: 2708-7980, online ISSN: 2708-7999
 Copyright © Asia Pac J Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Asia Pac J Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript