Review | Open Access
Liquid biopsy: an emerging field with new opportunities for cancer diagnosis and prognosis
Dhahiri Saidi Mashausi1, Semukunzi Herve2
1University of Dar es salaam College of Natural and Applied Sciences, Dar es Salaam, Tanzania.
2University of Rwanda, College of Medicine and Health Sciences, School of Medicine and Pharmacy, Department of Pharmacology and Toxicology, Kigali, Rwanda.
Correspondence: Semukunzi Herve (University of Rwanda, College of Medicine and Health Sciences, School of Medicine and Pharmacy, Department of Pharmacology and Toxicology, Kigali, Rwanda; E-mail: semukunziherve@gmail.com).
Asia-Pacific Journal of Oncology 2025, 6: 9-17. https://doi.org/10.32948/ajo.2025.01.09
Received: 08 Dec 2024 | Accepted: 09 Jan 2025 | Published online: 14 Jan 2025
Key words cancer, liquid biopsy, circulating tumor cells, circulating tumor DNA, exosomes, tumor educate platelets, miRNAs, lncRNAs, circRNAs
These tests are essentially non-invasive for patients, highlighting the potential of liquid biopsies to identify and continuously track tumor development [5]. Liquid biopsy has relied on entities like circulating tumor cells (CTCs) [6], circulating tumor DNA (ctDNA) [7], tumor-derived exosomes [8], tumor-educated platelets (TEPs)[9], and non-coding RNAs (ncRNA) [10]. Modern investigations focus chiefly on pinpointing CTCs, ctDNA, exosomes, TEPs and ncRNAs as shown in Figure 1. In this article we specifically offer an in-depth examination of different markers with prospects for deployment in liquid biopsy. Alongside, the present-day uses of liquid biopsy across miscellaneous cancer types were also discussed.
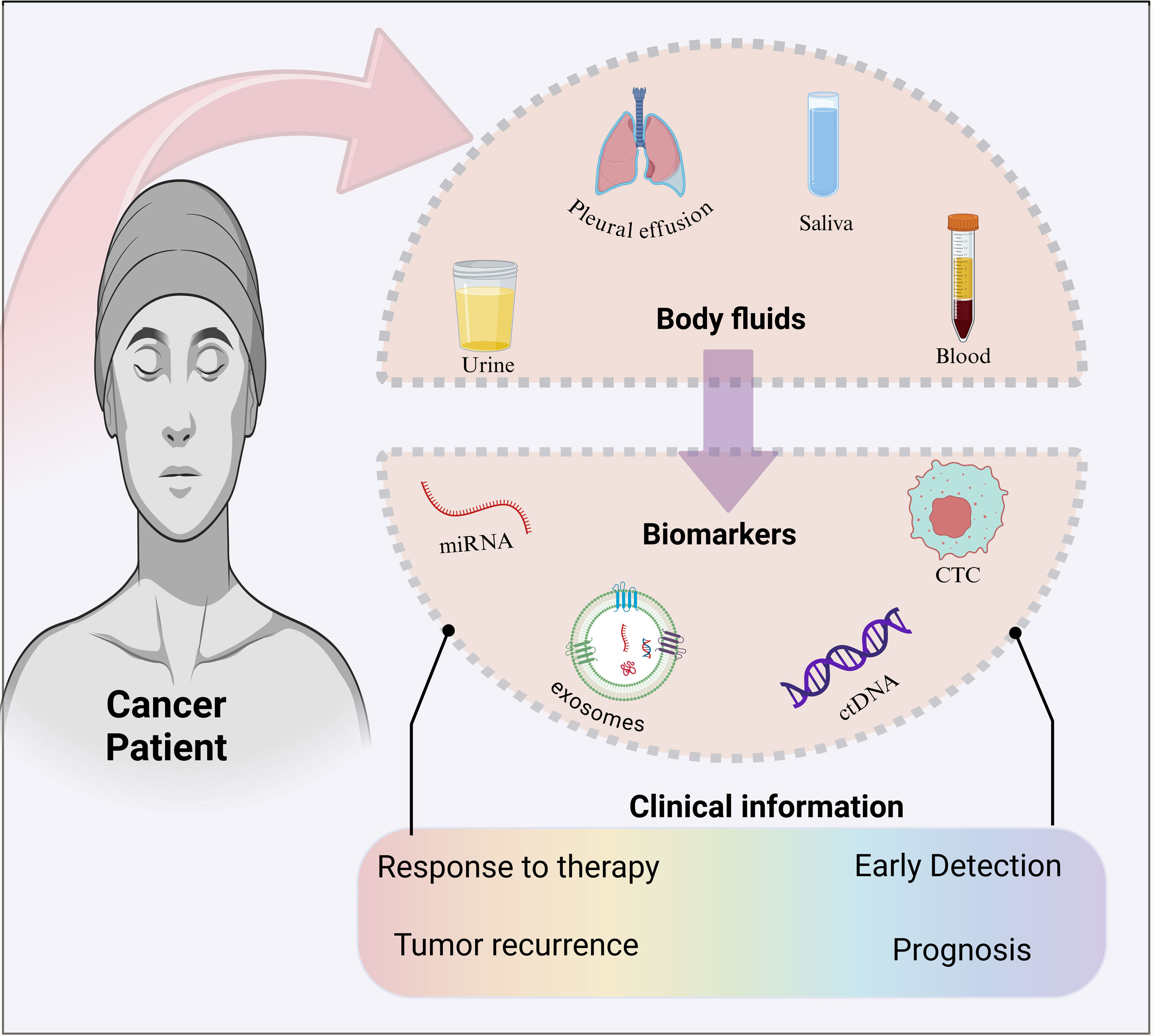 Figure 1. Schematic representation of the liquid biopsy technology. The most common body fluids and the most common markers that are deployed in liquid biopsy are represented. Meanwhile, liquid biopsy provides information that can be indispensable for clinical outcomes. Figure created with Biorender resources (Biorender.com).
Figure 1. Schematic representation of the liquid biopsy technology. The most common body fluids and the most common markers that are deployed in liquid biopsy are represented. Meanwhile, liquid biopsy provides information that can be indispensable for clinical outcomes. Figure created with Biorender resources (Biorender.com).
During the age of scientific advancement, scientists successfully isolated CTCs from blood samples for the first time in 1998, demonstrating a connection to pathological staging. This discovery paved the way for CTC utilization in clinical settings [17]. Before this, scientists in 1994 leveraged PCR techniques to recognize the initial KRAS mutation cfDNA extracted from patients of pancreatic cancer, obtained results that parallels those obtained in tumor tissue samples [18]. Raposo conducted a study that provided support for the biological function of EVs. Subsequent investigations have revealed that EVs derived from cells of the immune system can serve as presenters of antigens [19]. CTCs quantity prior to treatment acts as an independent marker for both overall survival and disease-free interval in patients with advanced breast cancer [20]. The ensuing expansion of the industry led to the inclusion of diverse liquid biopsy markers in oncology guidelines and their endorsement for clinical use [21].
Circulating tumor cells
A notable breakthrough in the research of CTCs happened in 1869 when Ashworth et al. discovered these cells in the blood of deceased cancer patients. CTCs, which stem from both primary and metastatic tumors, enter the circulatory or lymphatic systems of individuals with cancer and travel through their peripheral blood [23]. Despite their rarity, with only one CTC per million leukocytes and a short lifespan of 1-2.5 hours, recent research has shown a link between CTC concentrations and cancer advancement, especially in metastasis. CTCs are potent diagnostic maker of cancer and can be deployed for collecting important data for both clinical and scientific purposes [24]. Although improving the accuracy of CTC isolation and collection methods remains an ongoing issue, recent sophistication in technology have made their practical use in clinical environments possible. Technological progress has enhanced the precision in correlating CTC quantities with cancer development [25]. Escalation in the level of CTCs can be indictive of shorter progression-free survival and overall survival times. Escalated CTCs level in blood of breast cancer patient can be linked to short period of progression-free survival [26]. Consequently, CTCs detection can be key liquid biopsy marker [27].
The minuscule quantity of CTCs necessitates the use of highly sophisticated and sensitive methods for their effective isolation and detection. These techniques are constantly evolving, becoming more refined and accurate. Traditional methods leverage physical properties such as size and deformability, using techniques such as density gradient centrifugation, inertial focusing, and filtration. Some methods rely on CTCs specific markers, including epithelial cell adhesion molecules (EpCAM), vimentin, and N-cadherin [28]. Other strategies deploy marker specific immune-enrichment or immunomagnetic extraction, and used tools relying on microfluidics. To date the only FDA- authorized platform for CTCs quantification in blood is CellSearch® [29]. Despite their limitations, these methods have contributed greatly in furthering research on CTCs deployment for clinical purposes. As a minimally invasive diagnostic tool, CTCs are expected to become increasingly important in tumor early identification, progression and treatment intervention [30].
Circulating tumor DNA
Similar to CTCs, ctDNA can be isolated from blood and other bodily fluids such as ascites, pleural fluid, urine, and cerebrospinal fluid (CSF). cfDNA majorly stems from typical white blood cells and stromal cells [31]. Unlike cfDNA, ctDNA represents the existing state of the tumor. Analysis has confirmed that in cancer patients, ctDNA fragments are typically 20-50 base pairs shorter than cfDNA [32]. This characteristic renders ctDNA less vulnerable to the impact of heterogeneity within tumors. Furthermore, the brief half-life of ctDNA is crucial for its application as a real-time tumor indicator. These features provide ctDNA a marked benefit over conventional biopsy markers. In recent times, scientists have recognized the significance of ctDNA diagnostic and prognostic abilities for cancer [33]. Serum analysis of individuals with pancreatic cancer indicated increased concentrations of ctDNA, that diminished upon treatment, a marked benefit for deployment as a monitoring tool for treatment response [34].
Several existing clinical applications concentrate on detecting mutations in particular genes within ctDNA. Analysis of serum ctDNA of 18 colorectal cancer patients’ uncovered hotspot mutations in genes like APC, KRAS, TP53, and PIK3CA. ctDNA mutation can be indictive of patient response to treatment. Genetic modifications can disrupt the equilibrium of oncogenes, potentially leading to the onset of cancer. Therefore, detecting mutations in ctDNA is indispensable for cancer identification [35]. With the improvement is technology many robust techniques have been created for detecting ctDNA; such as RT-qPCR, ddPCR, and several standard and cutting-edge sequencing platforms [36]. It is anticipated that ctDNA tests will be extensively used in future for clinical and research purposes in the context of cancer theragnostic.
Exosomes
Exosome were first spotted in sheep reticulocytes. These structures are classified as a type of extracellular vesicles originating from endosomes. Exosomes form through membrane budding within multivesicular bodies and are released when these bodies merge with the cell membrane. Along with microvesicles and apoptotic vesicles, exosomes comprise the three main types of extracellular vesicles, primarily differentiated by their size and cellular origins [37]. Recent years have witnessed growing interest in these three subtypes. Exosomes can be detected in diverse bodily fluids, including blood, saliva, and urine. These structures fulfill various biological functions, encompassing molecular transportation, cellular communication, and immunological reactions of the body [38].
Exosomes are key player in the environment surrounding the tumor. Their impactful role in cancer progression is widely recognized [39]. The remarkable stability and comprehensive tumor cell data representation of exosomes significantly enhance liquid biopsy applications [40]. Recent cancer research has extensively explored the various exosomal components, including nucleic acids, proteins, lipids, and metabolites. Exosomal non-coding RNAs (ncRNAs) have emerged as particularly promising cancer diagnosis and treatment indicators [41]. Studies have revealed that heightened levels of specific exosomal miRNAs, [42] such as “miR-1246, miR-4644, miR-3976 and miR-4306” can function as highly sensitive prostate cancer biomarkers [43]. Serum form the patients of bladder cancer have escalated level of “exosomal lncRNA H19” , pointing to the possibility that exosomal lncRNAs can be used as reliable biomarkers for its diagnosis [44].
The surge in research interest surrounding exosomal proteins can be attributed to their extensive diversity and high concentrations. These proteins are known to play pivotal roles in modifying the cancer microenvironment, fostering tumor expansion, and facilitating cancer dissemination [45]. Furthermore, exosomal proteins have been associated with the emergence of chemotherapy resistance in individuals battling cancer. A recent study has highlighted the role of plasma gelsolin (pGSN), a variant of the GSN protein secreted by chemotherapy-resistant ovarian cancer cells. Exosomes can facilitate the transport of this protein, which subsequently activates α5β1 integrin, resulting in elevated levels of hypoxia-inducible factor 1 subunit α. As a result, this activation enhances ovarian cancer cells prowess to withstand chemotherapy and survive [46].
The crucial role of exosomes as key markers in liquid biopsies, along with their significant clinical importance, highlights the need for robust and precise methods for their extraction and identification [47]. Recently, analytical techniques such as RT-PCR, genome sequencing, and proteomics have become more accessible for analyzing exosomal contents [48]. Commonly used methods for exosomes extraction include differential ultracentrifugation, size-based separation, immunomagnetic isolation, and microfluidic approaches. As technology advances and various scientific fields converge, it is expected that exosomes incorporated liquid biopsies would make it to clinical practice expeditiously, particularly in the realm of cancer detection [49].
Tumor educated-platelets
Originally associated primarily with blood clotting and thrombosis, platelets are now seen as important in cancer. Ranking as the second most numerous cell type in blood, platelets are vital to numerous physiological functions [50]. These encompass aiding in wound healing, involvement in development of atherosclerosis, regulating vascular growth, and affecting the process of formation of new blood vessels. A significant connection has been revealed between increased platelet levels and cancer, subsequent research reinforced this connection [51]. Studies have shown a positive correlation between platelet accumulation and mortality in cancer patients. Recently, a specific type of platelet known as TEPs have emerged as a new liquid biopsy candidate. TEPs are obtained from cancer patients and exhibit distinct RNA and protein profiles. Research suggests that TEPs assist in the growth and spread of various solid tumors [52]. In particular, spliced TEP RNA markers can yield precise details about the existence, site, and molecular characteristics of tumors, even so, the detailed mechanisms call for further examination [53]. TEPs have not yet made to clinical practice, however, several studies have indicated TEPs' diagnostic potential, anticipating their indispensable role.
Platelets and tumors exert a mutual influence on each other. Platelets continuously incorporate tumor-derived components such as proteins, nucleic acids, vesicles, and granules that alters their RNA and protein expression profiles. As components of liquid biopsies, platelets present numerous benefits; exhibit notable stability and can be easily isolated through simple centrifugation at low speeds. The genetic content found in platelets demonstrates remarkable resilience. TEP short life allows for an accurate representation of the tumor’s current condition, facilitating ongoing surveillance in real time [52]. Recent research on platelets in individuals with tumors has largely focused on mRNA and lncRNAs. From the analysis of RNA sequencing data cancer patient can be successfully discerned from healthy subjects. Four specific lncRNA markers such as “LNCAROD, SNHG20, LINC00534, and TSPOAP-ASI” in platelets associated with colorectal cancer showed escalated expression both in platelets and serum samples from CRC patients, calling for lncRNAs diagnostic potential [54]. Establishment of a gene expression database specifically for platelet-based disease studies, would expedite the research progress aiming at liquid biopsies employing platelets. Nevertheless, our current grasp of the mechanisms underlying platelet RNA remains incomplete. TEPs deployment in cancer therapy is still at a conceptual stage, mandating extensive additional examination [55].
Non-coding RNAs
Unlike coding RNAs, non-coding RNAs have diverse functions within cells. Once dismissed as "junk RNA" and deemed irrelevant to cancer progression, modern research recognizes a fundamental role of non-coding RNAs in cancer onset and progression. Liquid biopsy deploying certain non-coding RNAs had shown excellent sensitivities and prospects for cancer detection [56].
miRNA and lncRNAs. MicroRNAs (miRNAs) are single-stranded RNA sequences of about “18 to 23” nucleotides, one of the categories of non-coding RNA. Have a recognized function in regulating gene expression post-transcriptionally. They attach to particular locations in the mRNA's 3′ untranslated region, diminishes the mRNA stability and inhibiting gene expression [57]. In cancer research, particularly within the realm of liquid biopsies, miRNAs are most thoroughly investigated type of non-coding RNAs. miR-21 and miR-155 escalated level are reported as indictive markers for numerous cancers, highlighting their prospective deployment as reliable liquid biopsy markers [58]. Recently, researchers have developed various miRNA detection methods, including qPCR, hybridization chain reaction, rolling circle amplification, and strand displacement amplification. These methods have played a crucial role in advancing miRNA studies, especially in elucidating its fundamental characteristics: prevalence and stability within tissues [59].
In cancer liquid biopsies, long non-coding RNAs (lncRNAs) are the second most studied non-coding RNAs. These RNA molecules, which exceed “200 nucleotides” in length and lack protein-coding capacity, perform various biological functions. They regulate gene transcription, influence miRNA control of target genes, and directly engage with proteins to influence their function and stability. Some lncRNAs also play roles in cell cycle control, impacting cellular growth and differentiation [60]. lncRNAs play critical role in cancer progression by regulating key cancer-associated transcriptional activators. There is indication of lncRNA involvement in tumor heterogeneity as there exists specific link between its expression and the type of tissue. Numerous cancer-associated lncRNAs levels are upraised in cancer patients derived serum and plasma [61]. For instance, in pancreatic ductal adenocarcinoma (PDAC) lncRNA have been shown to be useful as indictive liquid biopsy marker for the disease [62]. Upraised levels of lncRNA H19 are noticed in the plasma obtained from lung cancer patients, that heightened their possibility to be deployed as supplementary biomarker for lung cancer diagnosis [63]. As new lncRNAs are discovered expeditiously, it would be beneficial to initiate investigations focusing on explaining their specific functions and impact in cancer. New investigations are needed to evaluate lncRNAs indispensability as liquid biopsy based diagnostic tool for diverse cancer types [64].
Currently, lncRNA based diagnostic and prognostic models are also active area of research [65]. One study deployed a rm6A immune-related lncRNA to create a risk model that can be used for robust prediction about bladder cancer prognosis, immune status, and treatment response [66]. A separate study created a cuproptosis-linked lncRNA profile using intersecting lncRNAs and successfully used it for forecasting hepatocellular carcinoma outcomes and assessing the efficacy of immune checkpoint blockade (ICB) treatment [67]. The success in clinical trial is fundamental to get approval for application in clinical setting, however, it is evident that the use of miRNA and lncRNA for biomarker modeling represents a major technological intervention in the field of liquid biopsy technology [68].
Circ-RNA. Circular RNAs (circRNAs) are unique RNA molecules with a closed-loop structure, unable to encode proteins. They were first detected in 1971 during research on potato spindle tuber disease and known as self-replicating, "virus-like" RNA with a minimal molecular weight. The closed loop structure and the absence of free 5′ and 3′ ends, make these RNAs resistant to nucleases [69]. Their closed loop structure was visualized using radioactive labeling. With the advancement in research on circRNAs , it has become more evident that circRNAs are widely present and perform fundamental functions in human cells and tissues, such as acting as microRNA sponges, influencing the splicing of precursor mRNA, enhancing transcription, altering their own stability and location through interactions with RNA-binding proteins (RBPs), and producing functional proteins [70]. CircRNAs lack a typical polyA tail, preventing their direct detection using polyA tail-dependent purification methods [69]. Researchers have deployed diverse methods to detect circRNAs, including RT-PCR, RNAseq, northern hybridization, and high-throughput sequencing. These techniques involve the creation of primers that target specific reverse splice sites. CircRNAs' resistance to RNA exonuclease degradation allows their enrichment by selectively eliminating linear RNA.
Depending on the associated pathways, circRNAs can function as either proto-oncogenes or oncogenes in cancer [71]. As an example, circHIPK3 enhances the proliferation and migration of cancer cells through its activation of the miR-124/STAT3 signaling pathway. It indirectly activates STAT3, a transcription factor linked to various oncogenes and cell proliferation, by inhibiting miR-124's suppressive effect on STAT3, thereby influencing tumor cell behavior. Circ-RNA ITCH functions as an oncogene in diverse cancer types. Circ-ITCH interacts with particular microRNAs (miR-7, miR-17, and miR-214), indirectly influencing the expression of target genes [72]. These microRNAs and their targets are potentially implicated in several tumor-associated signaling pathways, including the Wnt/β-catenin and PI3K/AKT cascades. Irregular expression of circ-ITCH may facilitate tumor development by disturbing the equilibrium of these pathways. Investigations have identified circ-ITCH as a downregulated oncogene in cancers of the ovaries, prostate, brain, and stomach [73]. Ultimately, circRNAs facilitate tumor progression through multiple mechanisms. These include stimulating cellular division, circumventing growth-limiting factors, augmenting invasive and metastatic capabilities, fostering new blood vessel growth, modifying cellular energy dynamics, and eliciting inflammatory processes [74].
 Figure 2. Comparison of the characteristics of various liquid biopsy markers. Figure created with Biorender resources (Biorendor.com).
Figure 2. Comparison of the characteristics of various liquid biopsy markers. Figure created with Biorender resources (Biorendor.com).
It is essential to realize that liquid biopsy in clinical environments is only representative of the state of that particular biomarker during the diseases, not the entire state of the diseases. As such, liquid biopsy cannot fully substitute for tissue biopsy; these techniques jointly provide a more comprehensive insight into tumor biology. For widespread deployment of liquid biopsy in healthcare settings, extensive efforts should be made in optimization and standardization of methods linked to these markers’ isolation, detection and other downstream analysis techniques. The progress and adoption of liquid biopsy technology could be accelerated through refining detection strategies, integrating liquid biopsy markers, or fusing liquid biopsy with complementary diagnostic tools. Despite some knowledge gaps, liquid biopsy has become a focal point of scientific inquiry and extensive research. Although challenges remain, this innovative approach shows significant potential for integration into clinical practice.
|
Table 1. List of studies implicating liquid biopsy biomarkers for diverse cancer type. |
|||||
|
Cancer |
Biomarker |
Source |
Level |
Utility |
Ref. |
|
LC |
ctDNA CTCs let-7i-3p, miR-154-5p miRNA |
Plasma Blood Serum |
Up Up Down |
Treatment response Early diagnosis Early diagnosis |
[75] [76] [77] |
|
HCC |
CTCs miR-221-3p, miR-223-3p, miR-10b5p, miR-21-5p |
Blood Plasma exosomes |
Up Up |
Early diagnosis Early diagnosis |
[78] [79] |
|
CRC |
miR-203 miR-21 ctDNA |
Serum exosome Plasma exosome plasma |
Up Up Up |
Prognosis Recurrence & Prognosis Treatment response |
[80] [81] [82] |
|
RCC |
ctDNA miR-328-3p miR-15a has-mir-92a-1-5p |
Plasma Urine Urine Plasma exosome |
Up Down Up Down |
Prognosis Prognosis Early diagnosis Early diagnosis |
[83] [84] [85] [86] |
|
UC |
ctDNA miR-141 miR-151b |
Plasma Serum Serum |
Up Up Up |
Treatment response Early diagnosis Prognosis |
[87] [88] [89] |
|
MM |
CTCs ctDNA ctDNA mutation |
Blood Plasma Serum |
Up Up Up |
Early diagnosis Progression Progression |
[90] [91] [92] |
|
TC |
CTCs ctDNA ctDNA methylation miR-29a |
Blood Plasma Serum Serum exosome |
Up Up Up Down |
Early diagnosis Treatment response Diagnoses & recurrence Diagnosis &prognosis |
[93] [94] [95] [96] |
|
Note: “LC: Lung cancer, HCC: Hepatocellular carcinoma, CRC: Colorectal cancer, RCC: Renal cell carcinoma, UC: Urological cancer, MM: Multiple myeloma, TC: Thyroid cancer, CTCs: circulating tumor cells, ctDNA: circulating tumor DNA”. |
|||||
No applicable.
Ethics approval
No applicable.
Data availability
The data will be available upon request.
Funding
None.
Authors’ contribution
DSM, SH contributed to the conception, design, writing of this review article, figures drawing and submitted the final version of the manuscript.
Competing interests
None.
- Han B, Zheng R, Zeng H, Wang S, Sun K, Chen R, Li L, Wei W, He J: Cancer incidence and mortality in China, 2022. J Natl Cancer Cent 2024, 4(1): 47-53.
- Hirahata T, Ul Quraish R, Quraish AU, Ul Quraish S, Naz M, Razzaq MA: Liquid Biopsy: A Distinctive Approach to the Diagnosis and Prognosis of Cancer. Cancer Inform 2022, 21: 11769351221076062.
- Febbo PG, Allo M, Alme EB, Cuyun Carter G, Dumanois R, Essig A, Kiernan E, Kubler CB, Martin N, Popescu MC, et al: Recommendations for the Equitable and Widespread Implementation of Liquid Biopsy for Cancer Care. JCO Precis Oncol 2024, 8: e2300382.
- Lone SN, Nisar S, Masoodi T, Singh M, Rizwan A, Hashem S, El-Rifai W, Bedognetti D, Batra SK, Haris M, et al: Liquid biopsy: a step closer to transform diagnosis, prognosis and future of cancer treatments. Mol Cancer 2022, 21(1): 79.
- Ring A, Nguyen-Sträuli BD, Wicki A, Aceto N: Biology, vulnerabilities and clinical applications of circulating tumour cells. Nat Rev Cancer 2023, 23(2): 95-111.
- Lin D, Shen L, Luo M, Zhang K, Li J, Yang Q, Zhu F, Zhou D, Zheng S, Chen Y, et al: Circulating tumor cells: biology and clinical significance. Signal Transduct Target Ther 2021, 6(1): 404.
- Tivey A, Church M, Rothwell D, Dive C, Cook N: Circulating tumour DNA - looking beyond the blood. Nat Rev Clin Oncol 2022, 19(9): 600-612.
- Yu D, Li Y, Wang M, Gu J, Xu W, Cai H, Fang X, Zhang X: Exosomes as a new frontier of cancer liquid biopsy. Mol Cancer 2022, 21(1): 56.
- Najafi S, Asemani Y, Majidpoor J, Mahmoudi R, Aghaei-Zarch SM, Mortezaee K: Tumor-educated platelets. Clin Chim Acta 2024, 552: 117690.
- Zhong P, Bai L, Hong M, Ouyang J, Wang R, Zhang X, Chen P: A Comprehensive Review on Circulating cfRNA in Plasma: Implications for Disease Diagnosis and Beyond. Diagnostics (Basel) 2024, 14(10): 1045.
- Alexandrou G, Mantikas KT, Allsopp R, Yapeter CA, Jahin M, Melnick T, Ali S, Coombes RC, Toumazou C, Shaw JA, et al: The Evolution of Affordable Technologies in Liquid Biopsy Diagnostics: The Key to Clinical Implementation. Cancers (Basel) 2023, 15(22): 5434.
- Rapanotti MC, Cenci T, Scioli MG, Cugini E, Anzillotti S, Savino L, Coletta D, Di Raimondo C, Campione E, Roselli M, et al: Circulating Tumor Cells: Origin, Role, Current Applications, and Future Perspectives for Personalized Medicine. Biomedicines 2024, 12(9): 2137.
- Lo YMD, Han DSC, Jiang P, Chiu RWK: Epigenetics, fragmentomics, and topology of cell-free DNA in liquid biopsies. Science 2021, 372(6538): eaaw3616.
- Wolf JM, Espadas J, Luque-Garcia J, Reynolds T, Casadevall A: Lipid Biosynthetic Genes Affect Candida albicans Extracellular Vesicle Morphology, Cargo, and Immunostimulatory Properties. Eukaryot Cell 2015, 14(8): 745-754.
- Hessvik NP, Llorente A: Current knowledge on exosome biogenesis and release. Cell Mol Life Sci 2018, 75(2): 193-208.
- Cisneros-Villanueva M, Hidalgo-Pérez L, Rios-Romero M, Cedro-Tanda A, Ruiz-Villavicencio CA, Page K, Hastings R, Fernandez-Garcia D, Allsopp R, Fonseca-Montaño MA, et al: Cell-free DNA analysis in current cancer clinical trials: a review. Br J Cancer 2022, 126(3): 391-400.
- Lianidou ES, Markou A, Strati A: The Role of CTCs as Tumor Biomarkers. Adv Exp Med Biol 2015, 867: 341-367.
- Pratt ED, Cowan RW, Manning SL, Qiao E, Cameron H, Schradle K, Simeone DM, Zhen DB: Multiplex Enrichment and Detection of Rare KRAS Mutations in Liquid Biopsy Samples using Digital Droplet Pre-Amplification. Anal Chem 2019, 91(12): 7516-7523.
- Raposo G, Stoorvogel W: Extracellular vesicles: exosomes, microvesicles, and friends. J Cell Biol 2013, 200(4): 373-383.
- Fabisiewicz A, Szostakowska-Rodzos M, Zaczek AJ, Grzybowska EA: Circulating Tumor Cells in Early and Advanced Breast Cancer; Biology and Prognostic Value. Int J Mol Sci 2020, 21(5): 1671.
- Domínguez-Vigil IG, Moreno-Martínez AK, Wang JY, Roehrl MHA, Barrera-Saldaña HA: The dawn of the liquid biopsy in the fight against cancer. Oncotarget 2018, 9(2): 2912-2922.
- Alix-Panabières C, Pantel K: Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov 2021, 11(4): 858-873.
- Alix-Panabières C, Schwarzenbach H, Pantel K: Circulating tumor cells and circulating tumor DNA. Annu Rev Med 2012, 63: 199-215.
- Plaks V, Koopman CD, Werb Z: Cancer. Circulating tumor cells. Science 2013, 341(6151): 1186-1188.
- Paterlini-Brechot P, Benali NL: Circulating tumor cells (CTC) detection: clinical impact and future directions. Cancer Lett 2007, 253(2): 180-204.
- Cristofanilli M, Budd GT, Ellis MJ, Stopeck A, Matera J, Miller MC, Reuben JM, Doyle GV, Allard WJ, Terstappen LW, et al: Circulating tumor cells, disease progression, and survival in metastatic breast cancer. N Engl J Med 2004, 351(8): 781-791.
- Hong B, Zu Y: Detecting circulating tumor cells: current challenges and new trends. Theranostics 2013, 3(6): 377-394.
- de Wit S, van Dalum G, Lenferink AT, Tibbe AG, Hiltermann TJ, Groen HJ, van Rijn CJ, Terstappen LW: The detection of EpCAM(+) and EpCAM(-) circulating tumor cells. Sci Rep 2015, 5: 12270.
- Andree KC, van Dalum G, Terstappen LW: Challenges in circulating tumor cell detection by the CellSearch system. Mol Oncol 2016, 10(3): 395-407.
- Cortés-Hernández LE, Eslami SZ, Pantel K, Alix-Panabières C: Circulating Tumor Cells: From Basic to Translational Research. Clin Chem 2024, 70(1): 81-89.
- Mishra M, Ahmed R, Das DK, Pramanik DD, Dash SK, Pramanik A: Recent Advancements in the Application of Circulating Tumor DNA as Biomarkers for Early Detection of Cancers. ACS Biomater Sci Eng 2024, 10(8): 4740-4756.
- Dao J, Conway PJ, Subramani B, Meyyappan D, Russell S, Mahadevan D: Using cfDNA and ctDNA as Oncologic Markers: A Path to Clinical Validation. Int J Mol Sci 2023, 24(17): 13219.
- Pessoa LS, Heringer M, Ferrer VP: ctDNA as a cancer biomarker: A broad overview. Crit Rev Oncol Hematol 2020, 155: 103109.
- Guven DC, Sahin TK, Yildirim HC, Aktepe OH, Dizdar O, Yalcin S: A systematic review and meta-analysis of the association between circulating tumor DNA (ctDNA) and prognosis in pancreatic cancer. Crit Rev Oncol Hematol 2021, 168: 103528.
- Bach S, Sluiter NR, Beagan JJ, Mekke JM, Ket JCF, van Grieken NCT, Steenbergen RDM, Ylstra B, Kazemier G, Tuynman JB: Circulating Tumor DNA Analysis: Clinical Implications for Colorectal Cancer Patients. A Systematic Review. JNCI Cancer Spectr 2019, 3(3): pkz042.
- Ye P, Cai P, Xie J, Wei Y: The diagnostic accuracy of digital PCR, ARMS and NGS for detecting KRAS mutation in cell-free DNA of patients with colorectal cancer: A systematic review and meta-analysis. PLoS One 2021, 16(3): e0248775.
- Bastos N, Ruivo CF, da Silva S, Melo SA: Exosomes in cancer: Use them or target them? Semin Cell Dev Biol 2018, 78: 13-21.
- Zhu L, Sun HT, Wang S, Huang SL, Zheng Y, Wang CQ, Hu BY, Qin W, Zou TT, Fu Y, et al: Isolation and characterization of exosomes for cancer research. J Hematol Oncol 2020, 13(1): 152.
- Soung YH, Ford S, Zhang V, Chung J: Exosomes in Cancer Diagnostics. Cancers (Basel) 2017, 9(1): 8.
- Khan A, Raza F, He N: Nanoscale Extracellular Vesicle-Enabled Liquid Biopsy: Advances and Challenges for Lung Cancer Detection. Micromachines (Basel) 2024, 15(10): 1181.
- Eldh M, Lötvall J, Malmhäll C, Ekström K: Importance of RNA isolation methods for analysis of exosomal RNA: evaluation of different methods. Mol Immunol 2012, 50(4): 278-286.
- Yang B, Xiong WY, Hou HJ, Xu Q, Cai XL, Zeng TX, Ha XQ: Exosomal miRNAs as Biomarkers of Cancer: a Meta-Analysis. Clin Lab 2019, 65(5): p789.
- Madhavan B, Yue S, Galli U, Rana S, Gross W, Müller M, Giese NA, Kalthoff H, Becker T, Büchler MW, et al: Combined evaluation of a panel of protein and miRNA serum-exosome biomarkers for pancreatic cancer diagnosis increases sensitivity and specificity. Int J Cancer 2015, 136(11): 2616-2627.
- Ren J, Ding L, Zhang D, Shi G, Xu Q, Shen S, Wang Y, Wang T, Hou Y: Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics 2018, 8(14): 3932-3948.
- Li W, Li C, Zhou T, Liu X, Liu X, Li X, Chen D: Role of exosomal proteins in cancer diagnosis. Mol Cancer 2017, 16(1): 145.
- Beach A, Zhang HG, Ratajczak MZ, Kakar SS: Exosomes: an overview of biogenesis, composition and role in ovarian cancer. J Ovarian Res 2014, 7: 14.
- Huang R, Di K, Adeel K, Fan B, Gu X, Xu H, Shen H, He N, Li Z: The exploration of droplet digital branched rolling circle amplification based ultrasensitive biosensor for gastric cancer cell-derived extracellular vesicles detection. Mater. Today Adv 2022, 16: 100296.
- Xie H, Di K, Huang R, Khan A, Xia Y, Xu H, Liu C, Tan T, Tian X, Shen H: Extracellular vesicles based electrochemical biosensors for detection of cancer cells: A review. Chin. Chem. Lett 2020, 31(7): 1737-1745.
- Wang S, Khan A, Huang R, Ye S, Di K, Xiong T, Li Z: Recent advances in single extracellular vesicle detection methods. Biosens Bioelectron 2020, 154: 112056.
- Thon JN, Italiano JE: Platelets: production, morphology and ultrastructure. Handb Exp Pharmacol 2012, (210): 3-22.
- Wang Y, Zhang H, Li H, Xiong J, Wang J, Huang Y: Application of tumor-educated platelets as new fluid biopsy markers in various tumors. Clin Transl Oncol 2023, 25(1): 114-125.
- Plantureux L, Mège D, Crescence L, Dignat-George F, Dubois C, Panicot-Dubois L: Impacts of Cancer on Platelet Production, Activation and Education and Mechanisms of Cancer-Associated Thrombosis. Cancers (Basel) 2018, 10(11): 441.
- Heinhuis KM, In 't Veld S, Dwarshuis G, van den Broek D, Sol N, Best MG, Coevorden FV, Haas RL, Beijnen JH, van Houdt WJ, et al: RNA-Sequencing of Tumor-Educated Platelets, a Novel Biomarker for Blood-Based Sarcoma Diagnostics. Cancers (Basel) 2020, 12(6): 1372.
- Peterson JE, Zurakowski D, Italiano JE, Jr., Michel LV, Connors S, Oenick M, D'Amato RJ, Klement GL, Folkman J: VEGF, PF4 and PDGF are elevated in platelets of colorectal cancer patients. Angiogenesis 2012, 15(2): 265-273.
- Yu L, Guo Y, Chang Z, Zhang D, Zhang S, Pei H, Pang J, Zhao ZJ, Chen Y: Bidirectional Interaction Between Cancer Cells and Platelets Provides Potential Strategies for Cancer Therapies. Front Oncol 2021, 11: 764119.
- Yan H, Bu P: Non-coding RNA in cancer. Essays Biochem 2021, 65(4): 625-639.
- Xu D, Di K, Fan B, Wu J, Gu X, Sun Y, Khan A, Li P, Li Z: MicroRNAs in extracellular vesicles: Sorting mechanisms, diagnostic value, isolation, and detection technology. Front Bioeng Biotechnol 2022, 10: 948959.
- Yang M, Shen H, Qiu C, Ni Y, Wang L, Dong W, Liao Y, Du J: High expression of miR-21 and miR-155 predicts recurrence and unfavourable survival in non-small cell lung cancer. Eur J Cancer 2013, 49(3): 604-615.
- Khan A, Khan H, Mehwish F, Alam O, Khan MI, Ullah A, Atiq S, Ahmad M: MicroRNAs: The next generation of cancer biomarkers. Biomed Lett 2023, 9(2): 96-112.
- Mercer TR, Dinger ME, Mattick JS: Long non-coding RNAs: insights into functions. Nat Rev Genet 2009, 10(3): 155-159.
- Hauptman N, Glavač D: Long non-coding RNA in cancer. Int J Mol Sci 2013, 14(3): 4655-4669.
- Fu XL, Liu DJ, Yan TT, Yang JY, Yang MW, Li J, Huo YM, Liu W, Zhang JF, Hong J, et al: Analysis of long non-coding RNA expression profiles in pancreatic ductal adenocarcinoma. Sci Rep 2016, 6: 33535.
- Zhao Y, Feng C, Li Y, Ma Y, Cai R: LncRNA H19 promotes lung cancer proliferation and metastasis by inhibiting miR-200a function. Mol Cell Biochem 2019, 460(1-2): 1-8.
- Hashemi M, Moosavi MS, Abed HM, Dehghani M, Aalipour M, Heydari EA, Behroozaghdam M, Entezari M, Salimimoghadam S, Gunduz ES, et al: Long non-coding RNA (lncRNA) H19 in human cancer: From proliferation and metastasis to therapy. Pharmacol Res 2022, 184: 106418.
- Zhang Z, Luo K, Zou Z, Qiu M, Tian J, Sieh L, Shi H, Zou Y, Wang G, Morrison J, et al: Genetic analyses support the contribution of mRNA N(6)-methyladenosine (m(6)A) modification to human disease heritability. Nat Genet 2020, 52(9): 939-949.
- Hong W, Liang L, Gu Y, Qi Z, Qiu H, Yang X, Zeng W, Ma L, Xie J: Immune-Related lncRNA to Construct Novel Signature and Predict the Immune Landscape of Human Hepatocellular Carcinoma. Mol Ther Nucleic Acids 2020, 22: 937-947.
- Xu Q, Wang Y, Huang W: Identification of immune-related lncRNA signature for predicting immune checkpoint blockade and prognosis in hepatocellular carcinoma. Int Immunopharmacol 2021, 92: 107333.
- Li Y, Jiang T, Zhou W, Li J, Li X, Wang Q, Jin X, Yin J, Chen L, Zhang Y, et al: Pan-cancer characterization of immune-related lncRNAs identifies potential oncogenic biomarkers. Nat Commun 2020, 11(1): 1000.
- Ebbesen KK, Hansen TB, Kjems J: Insights into circular RNA biology. RNA Biol 2017, 14(8): 1035-1045.
- Nielsen AF, Bindereif A, Bozzoni I, Hanan M, Hansen TB, Irimia M, Kadener S, Kristensen LS, Legnini I, Morlando M, et al: Best practice standards for circular RNA research. Nat Methods 2022, 19(10): 1208-1220.
- Vo JN, Cieslik M, Zhang Y, Shukla S, Xiao L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X et al: The Landscape of Circular RNA in Cancer. Cell 2019, 176(4): 869-881.e813.
- Conn VM, Chinnaiyan AM, Conn SJ: Circular RNA in cancer. Nat Rev Cancer 2024, 24(9): 597-613.
- Su K, Yi Q, Dai X, Liu O: Circular RNA ITCH: An Emerging Multifunctional Regulator. Biomolecules 2022, 12(3): 359.
- Sun XD, Huan C, Sun DW, Lv GY: Prognostic and Clinicopathological Significance of Circular RNA circ-ITCH Expression in Cancer Patients: A Meta-analysis. Biomed Res Int 2021, 2021: 8828299.
- Cabanero M, Tsao MS: Circulating tumour DNA in EGFR-mutant non-small-cell lung cancer. Curr Oncol 2018, 25(Suppl 1): S38-S44.
- Zhang Z, Xiao Y, Zhao J, Chen M, Xu Y, Zhong W, Xing J, Wang M: Relationship between circulating tumour cell count and prognosis following chemotherapy in patients with advanced non-small-cell lung cancer. Respirology 2016, 21(3): 519-525.
- Huang J, Wu J, Li Y, Li X, Yang T, Yang Q, Jiang Y: Deregulation of serum microRNA expression is associated with cigarette smoking and lung cancer. Biomed Res Int 2014, 2014: 364316.
- Qi LN, Xiang BD, Wu FX, Ye JZ, Zhong JH, Wang YY, Chen YY, Chen ZS, Ma L, Chen J, et al: Circulating Tumor Cells Undergoing EMT Provide a Metric for Diagnosis and Prognosis of Patients with Hepatocellular Carcinoma. Cancer Res 2018, 78(16): 4731-4744.
- Ghosh S, Bhowmik S, Majumdar S, Goswami A, Chakraborty J, Gupta S, Aggarwal S, Ray S, Chatterjee R, Bhattacharyya S, et al: The exosome encapsulated microRNAs as circulating diagnostic marker for hepatocellular carcinoma with low alpha-fetoprotein. Int J Cancer 2020, 147(10): 2934-2947.
- Takano Y, Masuda T, Iinuma H, Yamaguchi R, Sato K, Tobo T, Hirata H, Kuroda Y, Nambara S, Hayashi N, et al: Circulating exosomal microRNA-203 is associated with metastasis possibly via inducing tumor-associated macrophages in colorectal cancer. Oncotarget 2017, 8(45): 78598-78613.
- Tsukamoto M, Iinuma H, Yagi T, Matsuda K, Hashiguchi Y: Circulating Exosomal MicroRNA-21 as a Biomarker in Each Tumor Stage of Colorectal Cancer. Oncology 2017, 92(6): 360-370.
- Osumi H, Shinozaki E, Yamaguchi K, Zembutsu H: Early change in circulating tumor DNA as a potential predictor of response to chemotherapy in patients with metastatic colorectal cancer. Sci Rep 2019, 9(1): 17358.
- Yamamoto Y, Uemura M, Fujita M, Maejima K, Koh Y, Matsushita M, Nakano K, Hayashi Y, Wang C, Ishizuya Y, et al: Clinical significance of the mutational landscape and fragmentation of circulating tumor DNA in renal cell carcinoma. Cancer Sci 2019, 110(2): 617-628.
- Di Meo A, Brown MD, Finelli A, Jewett MAS, Diamandis EP, Yousef GM: Prognostic urinary miRNAs for the assessment of small renal masses. Clin Biochem 2020, 75: 15-22.
- Mytsyk Y, Dosenko V, Borys Y, Kucher A, Gazdikova K, Busselberg D, Caprnda M, Kruzliak P, Farooqi AA, Lubov M: MicroRNA-15a expression measured in urine samples as a potential biomarker of renal cell carcinoma. Int Urol Nephrol 2018, 50(5): 851-859.
- Xiao CT, Lai WJ, Zhu WA, Wang H: MicroRNA Derived from Circulating Exosomes as Noninvasive Biomarkers for Diagnosing Renal Cell Carcinoma. Onco Targets Ther 2020, 13: 10765-10774.
- Blumendeller C, Boehme J, Frick M, Schulze M, Rinckleb A, Kyzirakos C, Kayser S, Kopp M, Kelkenberg S, Pieper N, et al: Use of plasma ctDNA as a potential biomarker for longitudinal monitoring of a patient with metastatic high-risk upper tract urothelial carcinoma receiving pembrolizumab and personalized neoepitope-derived multipeptide vaccinations: a case report. J Immunother Cancer 2021, 9(1): e001406.
- Kriebel S, Schmidt D, Holdenrieder S, Goltz D, Kristiansen G, Moritz R, Fisang C, Müller SC, Ellinger J: Analysis of tissue and serum microRNA expression in patients with upper urinary tract urothelial cancer. PLoS One 2015, 10(1): e0117284.
- Montalbo R, Izquierdo L, Ingelmo-Torres M, Lozano JJ, Capitán D, Alcaraz A, Mengual L: Prognostic value of circulating microRNAs in upper tract urinary carcinoma. Oncotarget 2018, 9(24): 16691-16700.
- Garcés JJ, Bretones G, Burgos L, Valdes-Mas R, Puig N, Cedena MT, Alignani D, Rodriguez I, Puente D, Álvarez MG, et al: Circulating tumor cells for comprehensive and multiregional non-invasive genetic characterization of multiple myeloma. Leukemia 2020, 34(11): 3007-3018.
- Mithraprabhu S, Sirdesai S, Chen M, Khong T, Spencer A: Circulating Tumour DNA Analysis for Tumour Genome Characterisation and Monitoring Disease Burden in Extramedullary Multiple Myeloma. Int J Mol Sci 2018, 19(7): 1858.
- Rustad EH, Coward E, Skytøen ER, Misund K, Holien T, Standal T, Børset M, Beisvag V, Myklebost O, Meza-Zepeda LA, et al: Monitoring multiple myeloma by quantification of recurrent mutations in serum. Haematologica 2017, 102(7): 1266-1272.
- Sato T, Harao M, Nakano S, Jotsuka T, Suda N, Yamashita J: Circulating tumor cells detected by reverse transcription-polymerase chain reaction for carcinoembryonic antigen mRNA: distinguishing follicular thyroid carcinoma from adenoma. Surgery 2005, 137(5): 552-558.
- Ciampi R, Romei C, Ramone T, Matrone A, Prete A, Gambale C, Materazzi G, De Napoli L, Torregrossa L, Basolo F, et al: Pre- and Post-operative Circulating Tumoral DNA in Patients With Medullary Thyroid Carcinoma. J Clin Endocrinol Metab 2022, 107(8): e3420-e3427.
- Hu S, Ewertz M, Tufano RP, Brait M, Carvalho AL, Liu D, Tufaro AP, Basaria S, Cooper DS, Sidransky D, et al: Detection of serum deoxyribonucleic acid methylation markers: a novel diagnostic tool for thyroid cancer. J Clin Endocrinol Metab 2006, 91(1): 98-104.
- Wen Q, Wang Y, Li X, Jin X, Wang G: Decreased serum exosomal miR-29a expression and its clinical significance in papillary thyroid carcinoma. J Clin Lab Anal 2021, 35(1): e23560.
Asia-Pacific Journal of Oncology
print ISSN: 2708-7980, online ISSN: 2708-7999
 Copyright © Asia Pac J Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.
Copyright © Asia Pac J Oncol. This work is licensed under a Creative Commons Attribution-NonCommercial-No Derivatives 4.0 International (CC BY-NC-ND 4.0) License.

 Submit Manuscript
Submit Manuscript